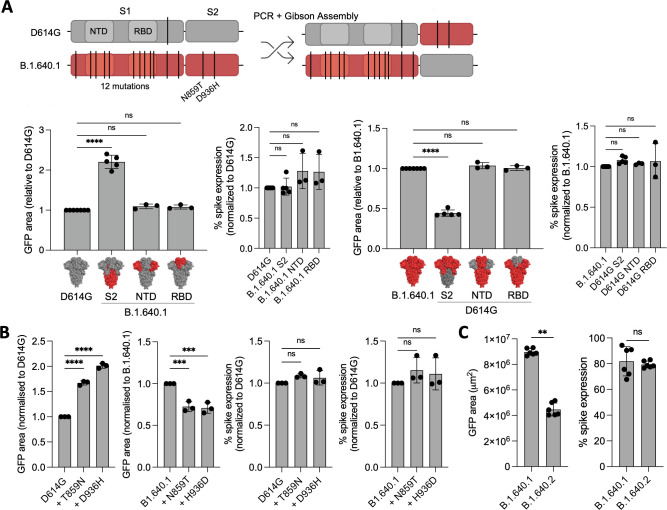
Fig 8.
B.1.640.1 Spike S2 mutations increase fusogenicity. (A) (Top) Schematic of D614G and B.1.640.1 chimeric Spike production through Gibson’s assembly. The NTD, RBD, and S2 domains of both Spikes were swapped to generate chimeric Spikes. For simplicity, only S1/S2 chimeras are shown. (Bottom) Fusion of D614G and B.1.640.1 Spike chimeras in HEK293T-GFP1-10 and VeroE6-GFP11 co-culture system. HEK293T-GFP1-10 cells were transfected with Spikes or control plasmids and then co-cultured with VeroE6-GFP11. Fusion was quantified by GFP area 18 hours post-transfection. Spike expression was assessed by surface staining of transfected HEK293T-GFP1-10 cells with an anti-S2 mAb. (B) D614G and B.1.640.1 Spikes were mutated to incorporate or revert B.1.640.1 S2-specific mutations, respectively. HEK293T-GFP1-10 cells were transfected with Spikes or control plasmids before co-culture with VeroE6-GFP11 cells. Fusion was assessed by GFP area 18 hours post-transfection. Spike expression was assessed by surface staining of transfected HEK293T-GFP1-10 cells with an anti-S2 mAb. (C) HEK293T-GFP1-10 cells were transfected with B.1.640.1 or B.1.640.2 Spikes or control plasmids before co-culture with VeroE6-GFP11 cells. Fusion was assessed by GFP area 18 hours post-transfection. Spike expression was assessed by surface staining of transfected HEK293T-GFP1-10 cells with an anti-S2 mAb. Mann-Whitney tests were performed to compare GFP area and spike expression, **P < 0.001. For (A) and (B), ordinary one-way ANOVA tests were performed with Tukey’s multiple comparison test to compare respective variants, **P < 0.001, ***P < 0.0001, ****P < 0.00001, ns = not significant. Error bars represent SD.