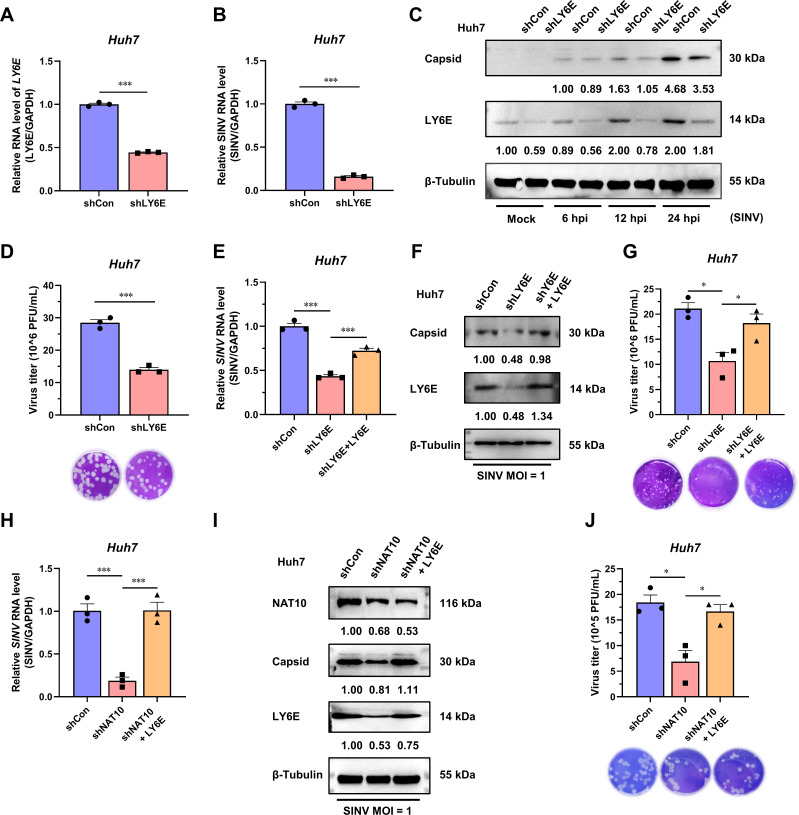
Fig 6.
SINV is positively affected by NAT10 as it regulates the stability of the LY6E mRNA. (A) qRT-PCR analysis of LY6E mRNA expression in LY6E-KD Huh7 cells. (B) qRT-PCR analysis of the SINV RNA expression levels in LY6E-KD Huh7 cells at 24 hpi (MOI = 1). (C) Immunoblot analysis of the SINV capsid protein expression in LY6E-KD Huh7 cells at 6, 12, and 24 hpi (MOI = 1). (D) Plaque formation assay using the SINV infectious virions obtained from the LY6E-KD Huh7 cell culture medium at 24 hpi (MOI = 1). (E) qRT-PCR analysis of the SINV RNA expression levels in LY6E-KD Huh7 cells ectopically expressing LY6E and infected with SINV, 24 hpi (MOI = 1). (F) Immunoblot analysis of the SINV capsid protein abundance described in panel (E). (G) Plaque formation assay using the SINV infectious virions obtained from the culture supernatant described in panel (E). (H) qRT-PCR analysis of the SINV RNA expression levels in NAT10-KD Huh7 cells ectopically expressing LY6E and infected with SINV, 24 hpi (MOI = 1). (I) Immunoblot analysis of the SINV capsid protein abundance as described in panel (H). (J) Plaque formation assay for the SINV infectious virions obtained from the culture supernatant described in panel (H). Blots were quantified with ImageJ software and normalized to control levels. Data are presented as the means ± SEM (n = 3). *P ≤ 0.05 and ***P ≤ 0.001 (A, B, and D, unpaired Student’s t-tests; E, G, H, and J, one-way ANOVA with Tukey’s multiple comparisons test).