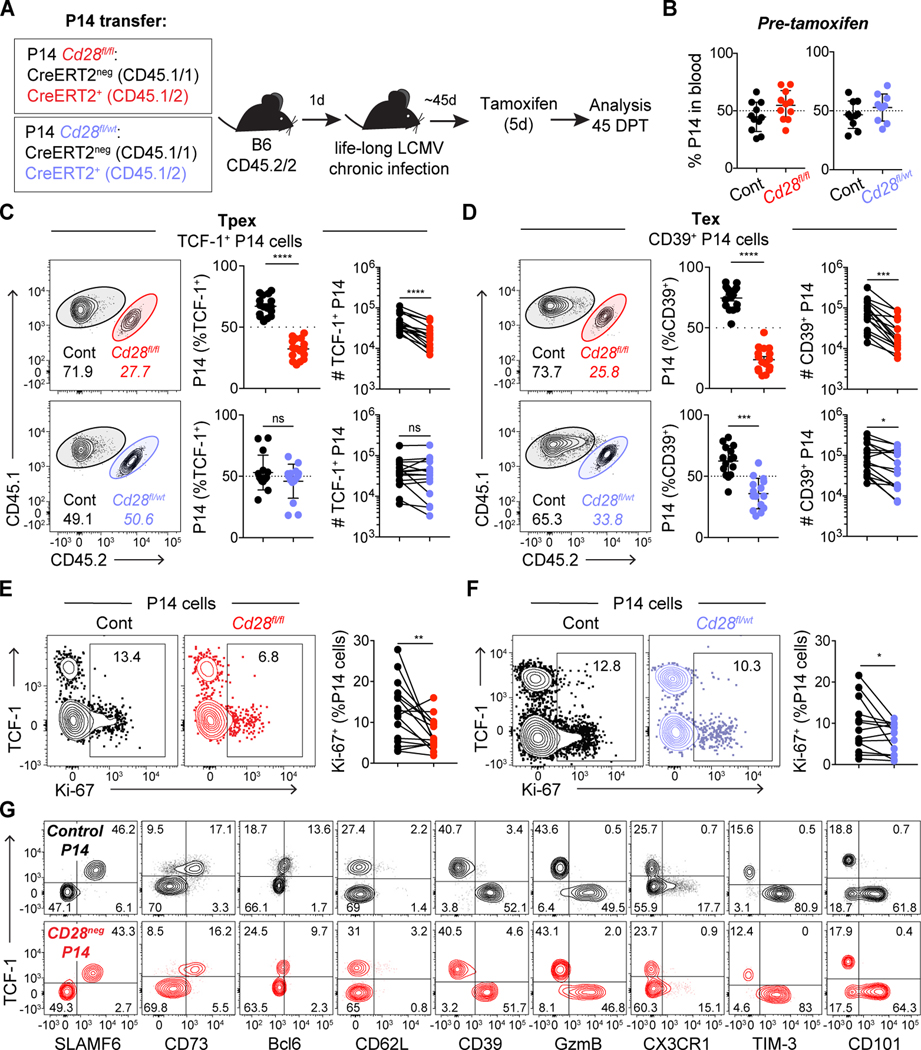
Fig. 2: Level of CD28 expression differentially impacts Tpex and Tex.
(A) Experimental layout: Cd28fl/fl CreERT2neg (control, black), Cd28fl/wt CreERT2+ (heterozygous, blue) and Cd28fl/fl CreERT2+ (homozygous, red) P14 T cells were co-transferred 50:50 (1,000 cells each) to C57BL/6J mice (depleted of CD4 T cells) the day before LCMV clone 13 infection. Mice that had been adoptively transferred with P14 T cells received tamoxifen 45 days post infection (DPI) and analyses in C-F were performed in spleen 45 days post tamoxifen (DPT). Analyses in C-F are on the entire population of transferred P14 T cells, without prior gating according to CD28 level. (B) Frequency of co-transferred P14 cells, 45 days post infection (DPI). (C) Frequency and absolute number of each P14 T cell population among Tpex (TCF-1+) and (D) Tex (CD39+). (E-F) Ki-67 expression. (G) Representative flow cytometry plots showing phenotypic characterization of P14 T cells: control (black) and Cd28fl/fl CreERT2+ gated on CD28neg (red) as shown in Fig S6A. Data in (B-G) are representative of 3 independent experiments with four to six mice per group. Data in (B) show combined data from two of three independent experiments; (C-F) show combined data of all three independent experiments. Symbols represent individual mice, bars show mean value of all animals analyzed and error bars indicate SEM. Connecting lines between symbols link cells analyzed from the same mouse. Significance in (C-F) was determined using a paired Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.