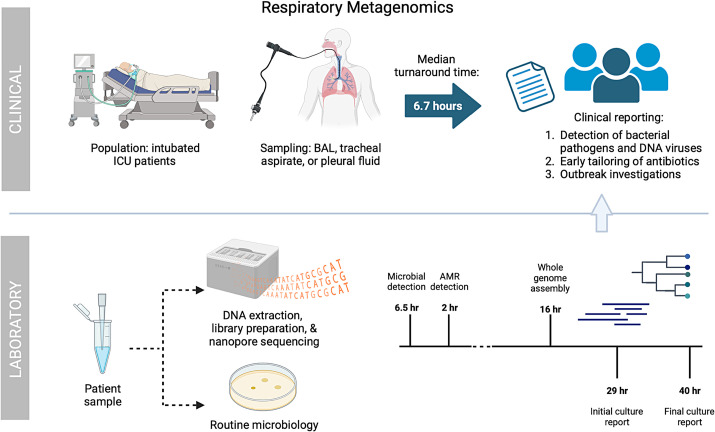
Figure 1.
Clinical implementation of respiratory metagenomics. Nanopore respiratory metagenomics were performed on BAL fluid, tracheal aspirates, and pleural fluid samples from intubated patients in the ICU with suspected respiratory infection. Routine clinical testing with culture was simultaneously performed. Sequencing reports were generated after 30 minutes and 2 hours for microbes and resistance mutations, respectively. Median turnaround time from sample acquisition to the reporting of results to clinicians was 6.7 hours. In comparison, preliminary culture results were reported after an average of 29 hours and were finalized in 40 hours. The impacts on clinical care, antibiotic agent prescribing, and infection control involvement were evaluated. AMR = antimicrobial resistance; BAL = bronchoalveolar lavage; ICU = intensive care unit.