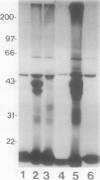
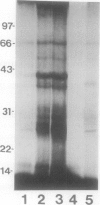
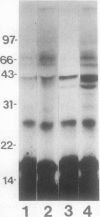
Abstract
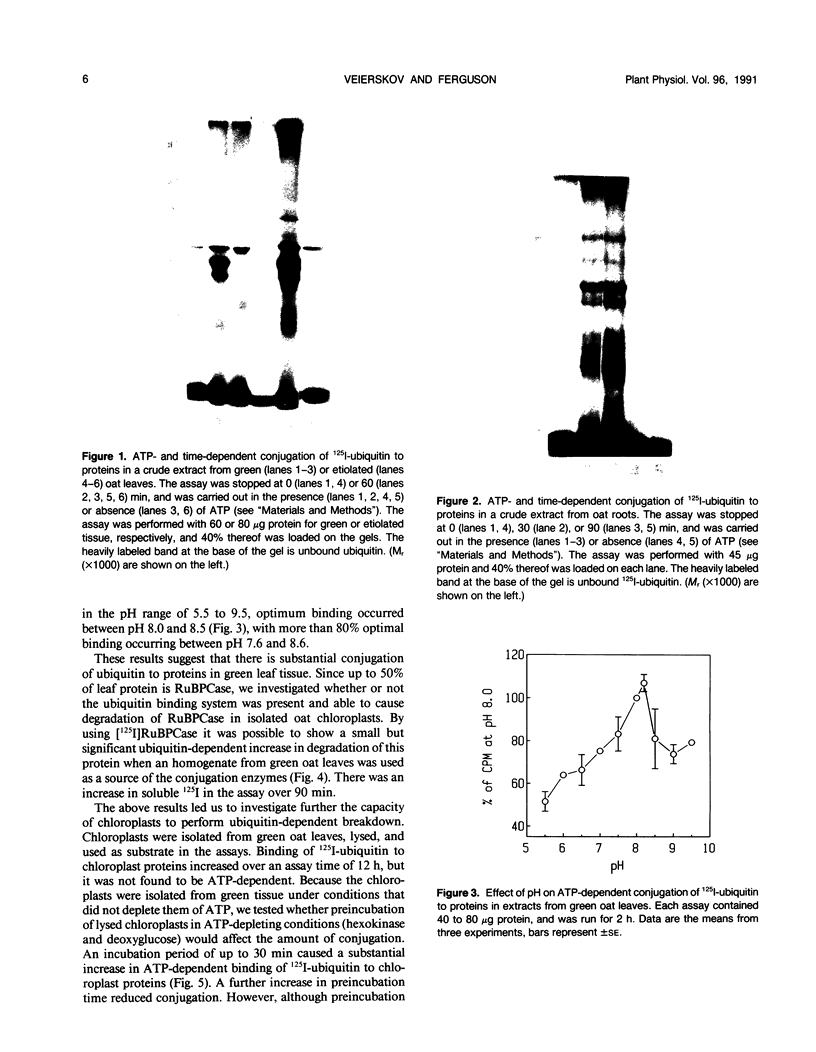
Conjugation of the polypeptide ubiquitin to endogenous proteins was studied in oat (Avena sativa L.) plants, and particularly in green tissues. Conjugating activity in leaf extracts was different from that in root extracts, and in both was less than in etiolated tissue. The conjugates were identified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and their formation was both time- and ATP-dependent and had a pH optimum of about 8.2. The assay had a high affinity for ATP with a probable Km of less than 50 micromolar. The ubiquitin conjugating system was also shown to be present in isolated chloroplasts, and ubiquitin could be conjugated to endogenous proteins of lyzed chloroplasts in which the ATP concentrations were reduced by preincubation or desalting. SDS-PAGE analysis led to the suggestion that the large and small subunits of ribulose-1,5-bisphosphate carboxylase (RuBPCase) may be able to be ubiquitinated, and we have shown that ubiquitin can stimulate the in vitro breakdown of 125I-labeled RuBPCase. These results invite the speculation that ubiquitin may be involved in the regulation of protein turnover in green plants.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ciechanover A., Heller H., Elias S., Haas A. L., Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1365–1368. doi: 10.1073/pnas.77.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganoth D., Leshinsky E., Eytan E., Hershko A. A multicomponent system that degrades proteins conjugated to ubiquitin. Resolution of factors and evidence for ATP-dependent complex formation. J Biol Chem. 1988 Sep 5;263(25):12412–12419. [PubMed] [Google Scholar]
- Haas A. L., Murphy K. E., Bright P. M. The inactivation of ubiquitin accounts for the inability to demonstrate ATP, ubiquitin-dependent proteolysis in liver extracts. J Biol Chem. 1985 Apr 25;260(8):4694–4703. [PubMed] [Google Scholar]
- Hammond J. B., Preiss J. ATP-Dependent Proteolytic Activity from Spinach Leaves. Plant Physiol. 1983 Dec;73(4):902–905. doi: 10.1104/pp.73.4.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. Mechanisms of intracellular protein breakdown. Annu Rev Biochem. 1982;51:335–364. doi: 10.1146/annurev.bi.51.070182.002003. [DOI] [PubMed] [Google Scholar]
- Hershko A., Leshinsky E., Ganoth D., Heller H. ATP-dependent degradation of ubiquitin-protein conjugates. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1619–1623. doi: 10.1073/pnas.81.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A. Ubiquitin-mediated protein degradation. J Biol Chem. 1988 Oct 25;263(30):15237–15240. [PubMed] [Google Scholar]
- Hough R., Pratt G., Rechsteiner M. Ubiquitin-lysozyme conjugates. Identification and characterization of an ATP-dependent protease from rabbit reticulocyte lysates. J Biol Chem. 1986 Feb 15;261(5):2400–2408. [PubMed] [Google Scholar]
- Jabben M., Shanklin J., Vierstra R. D. Ubiquitin-phytochrome conjugates. Pool dynamics during in vivo phytochrome degradation. J Biol Chem. 1989 Mar 25;264(9):4998–5005. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Musgrove J. E., Elderfield P. D., Robinson C. Endopeptidases in the stroma and thylakoids of pea chloroplasts. Plant Physiol. 1989 Aug;90(4):1616–1621. doi: 10.1104/pp.90.4.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport S., Dubiel W., Müller M. Proteolysis of mitochondria in reticulocytes during maturation is ubiquitin-dependent and is accompanied by a high rate of ATP hydrolysis. FEBS Lett. 1985 Jan 28;180(2):249–252. doi: 10.1016/0014-5793(85)81080-2. [DOI] [PubMed] [Google Scholar]
- Shanklin J., Jabben M., Vierstra R. D. Red light-induced formation of ubiquitin-phytochrome conjugates: Identification of possible intermediates of phytochrome degradation. Proc Natl Acad Sci U S A. 1987 Jan;84(2):359–363. doi: 10.1073/pnas.84.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer S. S., Choe H. T., Rausser S., Huffaker R. C. Characterization and subcellular localization of aminopeptidases in senescing barley leaves. Plant Physiol. 1988;87:894–897. doi: 10.1104/pp.87.4.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R. D. Demonstration of ATP-Dependent, Ubiquitin-Conjugating Activities in Higher Plants. Plant Physiol. 1987 Jun;84(2):332–336. doi: 10.1104/pp.84.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R. D., Langan S. M., Haas A. L. Purification and initial characterization of ubiquitin from the higher plant, Avena sativa. J Biol Chem. 1985 Oct 5;260(22):12015–12021. [PubMed] [Google Scholar]
- Vierstra R. D., Sullivan M. L. Hemin inhibits ubiquitin-dependent proteolysis in both a higher plant and yeast. Biochemistry. 1988 May 3;27(9):3290–3295. doi: 10.1021/bi00409a025. [DOI] [PubMed] [Google Scholar]
- Wettern M., Parag H. A., Pollmann L., Ohad I., Kulka R. G. Ubiquitin in Chlamydomonas reinhardii. Distribution in the cell and effect of heat shock and photoinhibition on its conjugate pattern. Eur J Biochem. 1990 Aug 17;191(3):571–576. doi: 10.1111/j.1432-1033.1990.tb19159.x. [DOI] [PubMed] [Google Scholar]
- van der Valk H. C., van Loon L. C. Subcellular Localization of Proteases in Developing Leaves of Oats (Avena sativa L.). Plant Physiol. 1988 Jun;87(2):536–541. doi: 10.1104/pp.87.2.536. [DOI] [PMC free article] [PubMed] [Google Scholar]