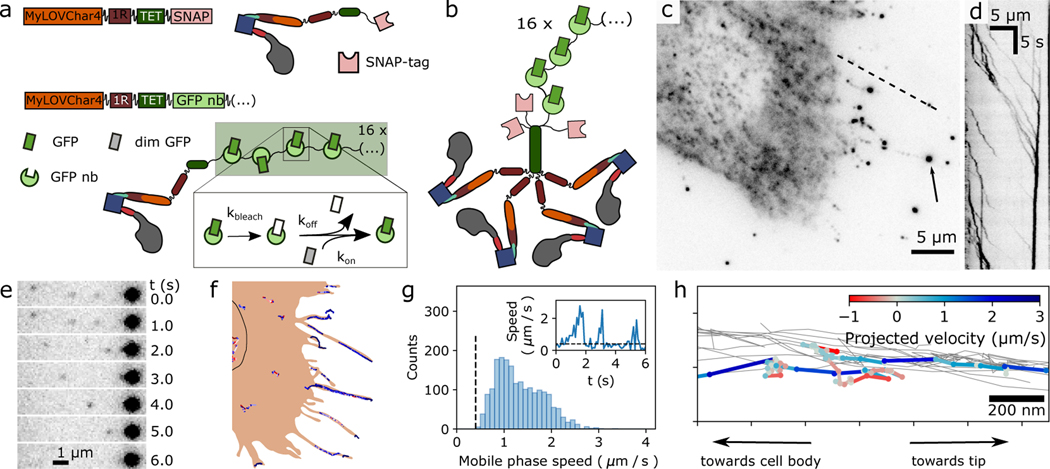
Figure 4. Single-molecule imaging of engineered motor constructs in live cells.
a, Design of molecular constructs for single-molecule tracking with high spatiotemporal resolution in cells. Block diagram and cartoons show two MyLOVChar4 constructs, C-terminally tagged with either a SNAP-tag (MyLOVChar4~1R~TET~SNAP) or a 16X ArrayG scaffold of GFP nanobodies (MyLOVChar4~1R~TET~ArrayG16X). Dim monomeric wildtype GFP (mwtGFP, grey rectangles) becomes brighter (green rectangles) upon binding to the scaffold; reversible association allows exchange of bleached mwtGFP (white rectangles) with unbleached protein from solution. b, Cartoon of the tetrameric motor complex imaged in cells overexpressing the non-visible species MyLOVChar4~1R~TET~SNAP, a minor population of MyLOVChar4~1R~TET~ArrayG16, and mwtGFP. c, Fluorescence image of a cell excited with 488 nm laser excitation. Arrow: bright fluorescence spot that is a terminal point of observed tracks. d, Kymograph along the dashed line in c. e-f, Single-molecule tracking of motors in cells. e, Image series from a region of the cell in panel c, with single motors moving toward, and merging with, a bright spot like the one indicated in panel c. f, Motor trajectories identified for the cell in panel c, overlaid on a schematic outline of the cell. Black outline, estimated location of the nucleus. Similar results to those presented in c-f were obtained on two independent biological replicates, across a total of 41 cells contained in 39 image acquisition series. g, Velocity histogram from all 41 cells, for all extracted directionally persistent tracks with a minimum length of 20 frames, tabulating mobile runs in which the speed exceeds 0.4 μm/s for 4 frames or longer (N= 1946 runs). The mean and mode of the distribution are 1.3 μm/s and 1.0 μm/s respectively. Inset: velocity trace with definition of mobile phases. h, Detail of motor trajectories in a cellular protrusion. Grey thin lines: set of all tracks associated with the protrusion. One selected trajectory is colored according to the projected velocity along the main axis of the protrusion.