Abstract
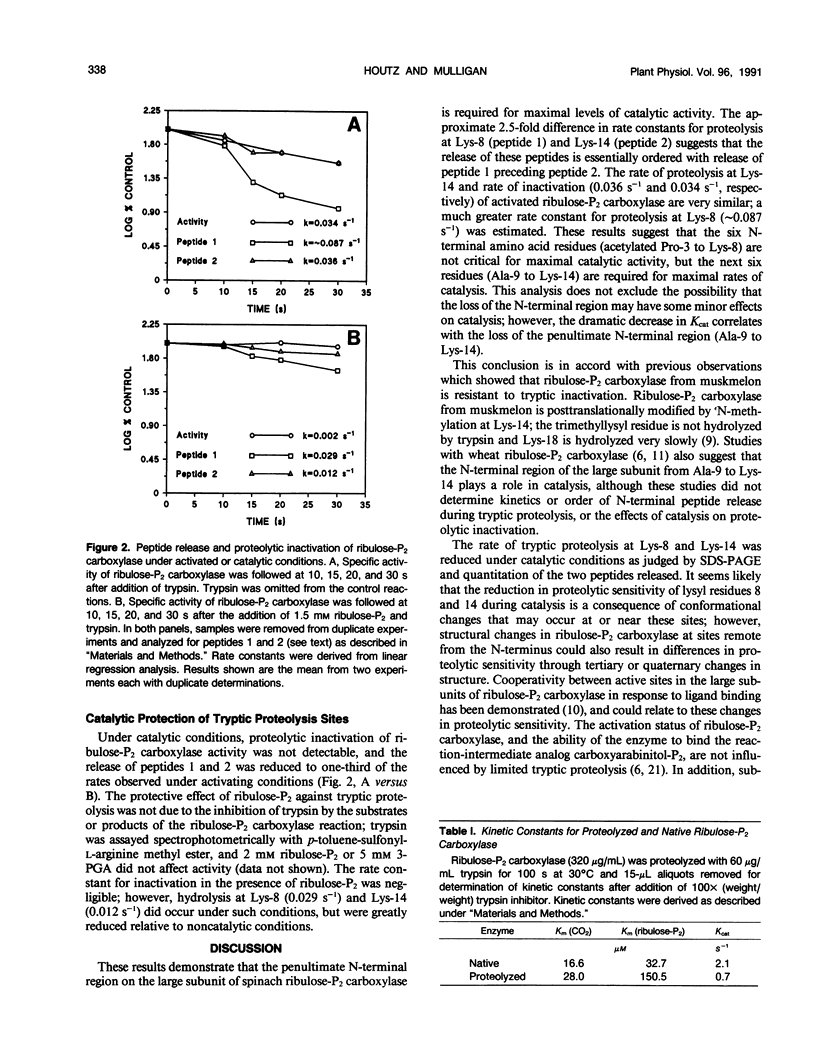
Limited tryptic proteolysis of spinach (Spinacia oleracea) ribulose bisphosphate carboxylase/oxygenase (ribulose-P2 carboxylase) resulted in the ordered release of two adjacent N-terminal peptides from the large subunit, and an irreversible, partial inactivation of catalysis. The two peptides were identified as the N-terminal tryptic peptide (acetylated Pro-3 to Lys-8) and the penultimate tryptic peptide (Ala-9 to Lys-14). Kinetic comparison of hydrolysis at Lys-8 and Lys-14, enzyme inactivation, and changes in the molecular weight of the large subunit, indicated that proteolysis at Lys-14 correlated with inactivation, while proteolysis at Lys-8 occurred much more rapidly. Thus, enzyme inactivation is primarily the result of proteolysis at Lys-14. Proteolysis of ribulose-P2 carboxylase under catalytic conditions (in the presence of CO2, Mg2+, and ribulose-P2) also resulted in ordered release of these tryptic peptides; however, the rate of proteolysis at lysyl residues 8 and 14 was reduced to approximately one-third of the rate of proteolysis of these lysyl residues under noncatalytic conditions (in the presence of CO2 and Mg2+ only). The protection of these lysyl residues from proteolysis under catalytic conditions could reflect conformational changes in the N-terminal domain of the large subunit which occur during the catalytic cycle.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chapman M. S., Suh S. W., Curmi P. M., Cascio D., Smith W. W., Eisenberg D. S. Tertiary structure of plant RuBisCO: domains and their contacts. Science. 1988 Jul 1;241(4861):71–74. doi: 10.1126/science.3133767. [DOI] [PubMed] [Google Scholar]
- Chen Z. X., Spreitzer R. J. Chloroplast intragenic suppression enhances the low CO2/O2 specificity of mutant ribulose-bisphosphate carboxylase/oxygenase. J Biol Chem. 1989 Feb 25;264(6):3051–3053. [PubMed] [Google Scholar]
- Houtz R. L., Stults J. T., Mulligan R. M., Tolbert N. E. Post-translational modifications in the large subunit of ribulose bisphosphate carboxylase/oxygenase. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1855–1859. doi: 10.1073/pnas.86.6.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johal S., Partridge B. E., Chollet R. Structural characterization and the determination of negative cooperativity in the tight binding of 2-carboxyarabinitol bisphosphate to higher plant ribulose bisphosphate carboxylase. J Biol Chem. 1985 Aug 15;260(17):9894–9904. [PubMed] [Google Scholar]
- Kettleborough C. A., Parry M. A., Burton S., Gutteridge S., Keys A. J., Phillips A. L. The role of the N-terminus of the large subunit of ribulose-bisphosphate carboxylase investigated by construction and expression of chimaeric genes. Eur J Biochem. 1987 Dec 30;170(1-2):335–342. doi: 10.1111/j.1432-1033.1987.tb13704.x. [DOI] [PubMed] [Google Scholar]
- Knight S., Andersson I., Brändén C. I. Crystallographic analysis of ribulose 1,5-bisphosphate carboxylase from spinach at 2.4 A resolution. Subunit interactions and active site. J Mol Biol. 1990 Sep 5;215(1):113–160. doi: 10.1016/S0022-2836(05)80100-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larimer F. W., Lee E. H., Mural R. J., Soper T. S., Hartman F. C. Intersubunit location of the active site of ribulose-bisphosphate carboxylase/oxygenase as determined by in vivo hybridization of site-directed mutants. J Biol Chem. 1987 Nov 15;262(32):15327–15329. [PubMed] [Google Scholar]
- Lorimer G. H. Ribulosebisphosphate carboxylase: amino acid sequence of a peptide bearing the activator carbon dioxide. Biochemistry. 1981 Mar 3;20(5):1236–1240. doi: 10.1021/bi00508a028. [DOI] [PubMed] [Google Scholar]
- Lundqvist T., Schneider G. Crystal structure of the complex of ribulose-1,5-bisphosphate carboxylase and a transition state analogue, 2-carboxy-D-arabinitol 1,5-bisphosphate. J Biol Chem. 1989 Apr 25;264(12):7078–7083. [PubMed] [Google Scholar]
- McCurry S. D., Gee R., Tolbert N. E. Ribulose-1,5-bisphosphate carboxylase/oxygenase from spinach, tomato, or tobacco leaves. Methods Enzymol. 1982;90(Pt E):515–521. doi: 10.1016/s0076-6879(82)90178-1. [DOI] [PubMed] [Google Scholar]
- Miziorko H. M., Lorimer G. H. Ribulose-1,5-bisphosphate carboxylase-oxygenase. Annu Rev Biochem. 1983;52:507–535. doi: 10.1146/annurev.bi.52.070183.002451. [DOI] [PubMed] [Google Scholar]
- Mulligan R. M., Houtz R. L., Tolbert N. E. Reaction-intermediate analogue binding by ribulose bisphosphate carboxylase/oxygenase causes specific changes in proteolytic sensitivity: the amino-terminal residue of the large subunit is acetylated proline. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1513–1517. doi: 10.1073/pnas.85.5.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce J. W., McCurry S. D., Mulligan R. M., Tolbert N. E. Activation and assay of ribulose-1,5-bisphosphate carboxylase/oxygenase. Methods Enzymol. 1982;89(Pt 500):47–55. doi: 10.1016/s0076-6879(82)89011-3. [DOI] [PubMed] [Google Scholar]
- Schneider G., Lindqvist Y., Brändén C. I., Lorimer G. Three-dimensional structure of ribulose-1,5-bisphosphate carboxylase/oxygenase from Rhodospirillum rubrum at 2.9 A resolution. EMBO J. 1986 Dec 20;5(13):3409–3415. doi: 10.1002/j.1460-2075.1986.tb04662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



