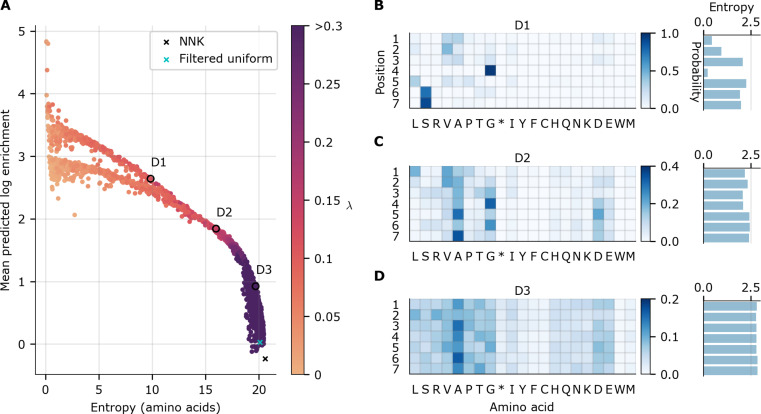
Fig. 4. Designed AAV5-based 7-mer insertion libraries.
Each point in (A) represents a theoretical library designed with our diversity-constrained optimal library approach, with one particular diversity constraint, λ (higher values yields more diverse libraries). Entropy indicates diversity of the library distribution, while MPLE indicates overall library fitness; both quantities were computed from the theoretical library distribution. The baseline NNK library is denoted with a black “×”, while a cyan “×” denotes the filtered uniform library that is uniform over all 21-mer nucleotide sequences except for those containing stop codons. Three designed libraries have been circled and labeled D1 to D3 for reference. Because of the non-convex optimization problem, some dots are suboptimal (i.e., lie strictly below or to the left of other dots) and are therefore further from the optimal frontier but are displayed for completeness. (B to D) Designed library parameters (probability of each amino acid at each position) for the three designed libraries D1 to D3, respectively, highlighted in (A).