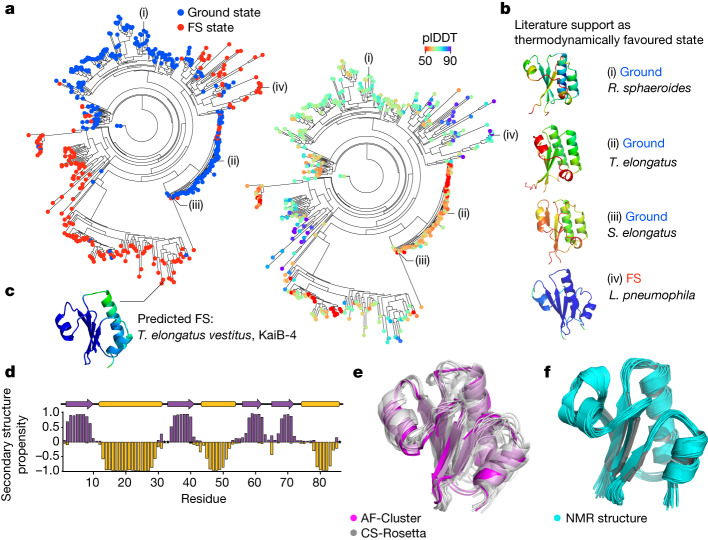
Fig. 2. The KaiB family contains pockets of sequences predicted to be stabilized for both states.
a, AF2 predictions for each variant in a phylogenetic tree using the ten closest sequences as the input MSA. Left, each node is coloured by predicted state (blue, ground state; red, FS state). Right, the same tree, coloured by plDDT. b, Three known fold-switching KaiB variants from R. sphaeroides32 (i), T. elongatus5 (ii) and S. elongatus31 (iii) are predicted in the ground state, and a variant from L. pneumonia37 (iv), crystallized in the FS state, is predicted in the FS state with a high plDDT. c, A KaiB copy present in T. elongatus vestitus, KaiBTV-4, is predicted to favour the FS state. d–f, Experimental testing of KaiBTV-4. d, The secondary structure propensity determined by NMR backbone chemical shifts, calculated using TALOS-N57 for KaiBTV-4, fully agrees with the FS state predicted by AF-Cluster. Unassigned amino acid residues are indicated by stars. e, Structure models calculated using CS-Rosetta39, shown in grey, have 1.8 ± 0.3 Å r.m.s.d. to the AF-Cluster model (magenta). f, NMR structural models calculated from 3D 1H-15N- and 3D 1H-13C-edited NOESY spectra have an average pairwise r.m.s.d. of 0.7 Å, and 1.89 ± 0.13 Å r.m.s.d. to the AF-Cluster model. r.m.s.d. values in e and f were calculated over backbone atoms in secondary structure regions.