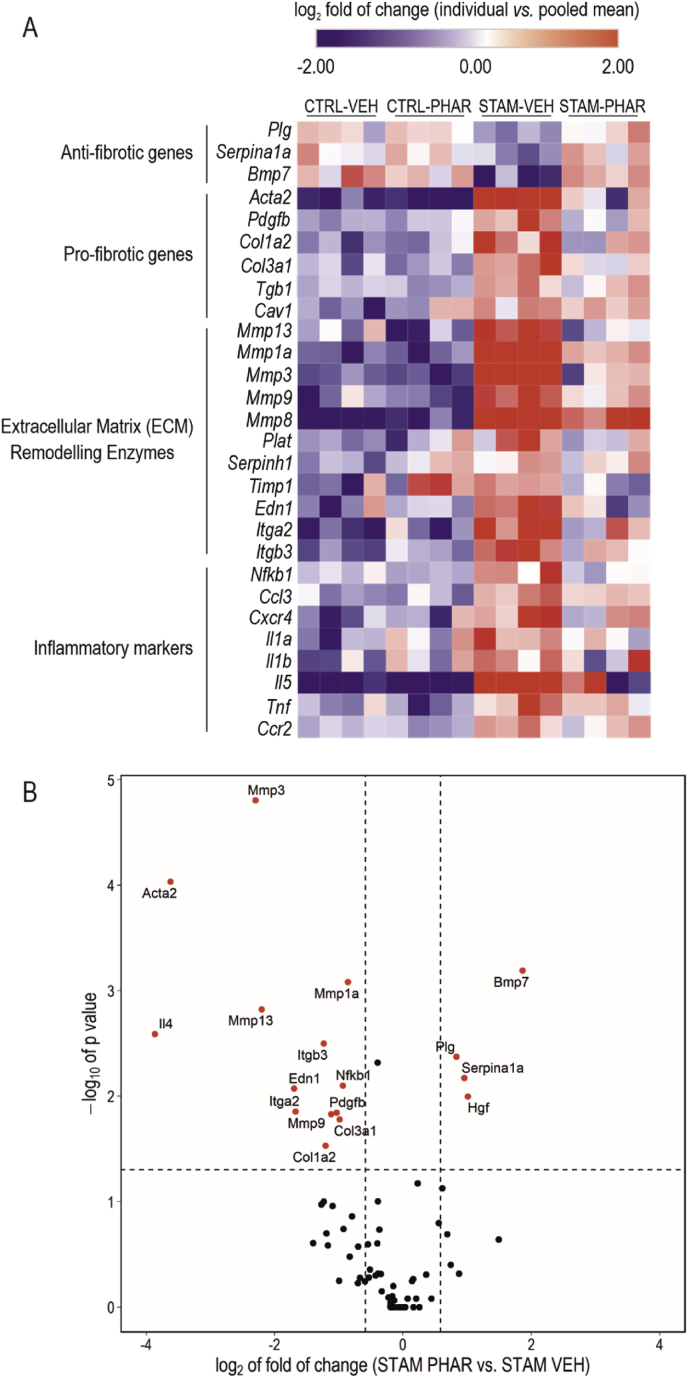
Fig. 5.
PHAR counteracts the fibrotic transcriptional signature. A, Changes in expression of 28 out of 84 genes involved in liver fibrosis were analyzed through RT2 Profiler™ PCR Array Mouse Fibrosis Qiagen for CTRL-VEH, CTRL-PHAR, STAM-VEH and STAM-PHAR livers. The 28 genes showed significant expression changes between CTRL-VEH vs. STAM-VEH (n = 4 per experimental group; p-value ≤0.05; two-way ANOVA, Bonferroni's post-hoc test). These genes were grouped by their biological function in a heatmap, representing the individual log2 of the fold of change. The color scale is capped at −2 and 2 of log2 fold of change. B, Volcano plot representing -log10 of p-value (calculated by a two way-ANOVA, Bonferroni's post-hoc test) and log2 of the fold of change for differences in gene expression between STAM-VEH and STAM-PHAR. The threshold p-value (dashed horizontal line) was established to <0.05, and the threshold for log2-fold change (dashed vertical lines) was established at -log2(1.5) and log2(1.5). Genes surpassing both thresholds are shown in red. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)