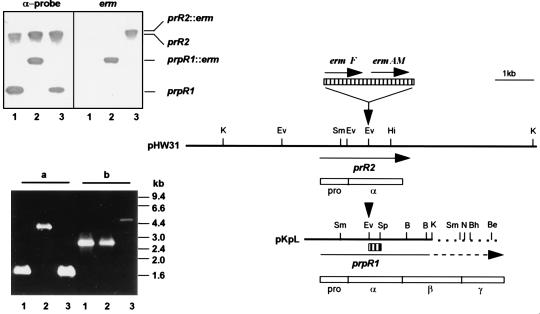
FIG. 1.
Genomic organization of the major arginine-specific protease genes prR2 and prpR1 and Southern and PCR analyses of P. gingivalis W50 strains. The two bold lines represent an ∼8.5-kb-HindIII (pHW31) or Sau3AI-KpnI (pKpL, nucleotide 1 to 3471 of the sequence with accession no. X82680) fragment of P. gingivalis W50 genomic DNA bearing prR2 and truncated prpR1, respectively. Unique restriction sites are shown as bars above the fragments. Note that both pKpL and prpR1 have been extended for comparative purposes, and this is shown as broken lines. Insertion of the Erm cassette, encoding ermF and ermAM (hatched box above restriction maps), is at the EcoRV site (indicated by down-pointing arrows) for generation of P. gingivalis W501 or W50D7 via electrotransformation. Positions and direction of genes are indicated by arrows under the DNA fragments. Empty boxes are schematic representations of translated sequences (PrRII and PrpRI) with their domain characteristics (pro, α, β, and γ); pro and α domains are common to the two gene products. The α-probe, used in the Southern hybridization experiment, is shown as a small hatched box under pKpL. In the Southern hybridization (upper inserted panel), P. gingivalis chromosomal DNA was restricted with SmaI and membranes were probed with either the 270-bp α-probe or the 2.1-kb Erm cassette. The positions of the 3.2-kb prpR1 and ≥12-kb prR2 together with the corresponding erm inserted loci are shown. Lanes: 1, W50 (wt); 2, W501 (prpR1 mutant); 3, W50D7 (prR2 mutant). The PCR panel (lower insert) shows the agarose electrophoresis of amplicons generated with primers specific for prpR1 (a) or prR2 (b) by using P. gingivalis chromosomal DNA as templates. The lane numbers are the same as for the Southern hybridization experiment. B, BamHI; Be, BstEII; Bh, BssHII; Ev, EcoRV; Hi, HincII; K, KpnI; N, NcoI; Sm, SmaI; Sp, SphI.