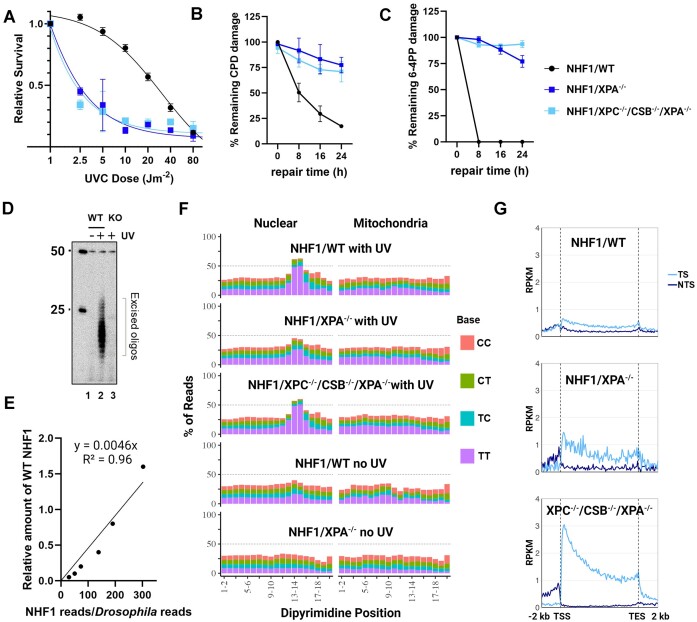
Figure 3.
Human NHF1/XPA−/− and NHF1/XPC−/−/CSB−/−/XPA−/− cells exhibit extreme UV sensitivity and nearly undetectable repair by low resolution assays yet demonstrate evidence of excision repair by qXR-Seq analysis. (A) The three NHF1 cell lines were analyzed for survival two days after the indicated doses of UVC. Shown are the mean from three biological replicates with error bars denoting standard error of the mean (SEM). Slot blot analysis showing the (B) CPD and (C) 6–4PP repair rates of the three NHF1 cell lines treated with 5 J/m2 UVC. Damaged-DNA signals were normalized to time = 0 and plotted as a function of time. Results shown are the mean from three biological replicates with error bars denoting SEM. (D) The in vivo excision assay was used to compare the amount of 6–4PP-containing excised oligos in extracts from the NHF1 wildtype (WT) and NHF1/ XPA−/− (KO) cell lines. An equal number of cells were either unirradiated (lane 1) or irradiated with 20 J/m2 UVC (lanes 2 and 3) and incubated 2h at 37°C to allow for repair. Cells were lysed by the Hirt procedure and low molecular weight DNA in the supernatant was immunoprecipitated with anti-6–4PP antibodies. The recovered oligos were mixed with a 50-mer internal control oligo, 5′-end labeled, and separated on a DNA sequencing gel along with the indicated size markers. (E) The qXR-seq method was used to map the genomic location of excision products in a quantitative manner. Excised oligos were isolated from human cells lysed with the Hirt method, mixed with excised oligos from Hirt-lysed UV-irradiated Drosophila cells, then purified with anti-(6–4)PP specific antibodies, ligated to adapters, and again purified with anti-(6–4)PP antibodies. The damage was reversed with (6–4)PP photolyase and PCR was performed to generate libraries for high throughput sequencing. The sequencing reads were uniquely mapped to either the human or fly genomes, and the graph shows the human:fly ratio relative to the amount of WT NHF1. (F) Analysis of the frequency of the possible dipyrimidines along CPD qXR-Seq reads of 24–30nt length (trimmed to 20nt from the 5′ end) from the indicated NHF1 cell lines mapped to either nuclear genome DNA (left) or mitochondrial DNA (right). Mitochondrial DNA analysis and unirradiated cells were included to control for specificity. (G) Analysis of transcription-coupled repair (TCR) in the indicated cell lines. CPD qXR-Seq data is plotted as average repair reads (y-axis) along the length of a ‘unit gene’ (x-axis) (5000 genes were selected and divided into 100 bins as described in Materials and methods. The median length for these 5000 genes is 26 196 bp). TCR can clearly be observed in NHF1/XPA−/− cells (middle) even though the plots are not as smooth due to low read numbers. RPKM, reads per kilobase per million mapped reads; TSS, transcription start site; TES, transcription end site; TS, transcribed strand; NTS, nontranscribed strand.