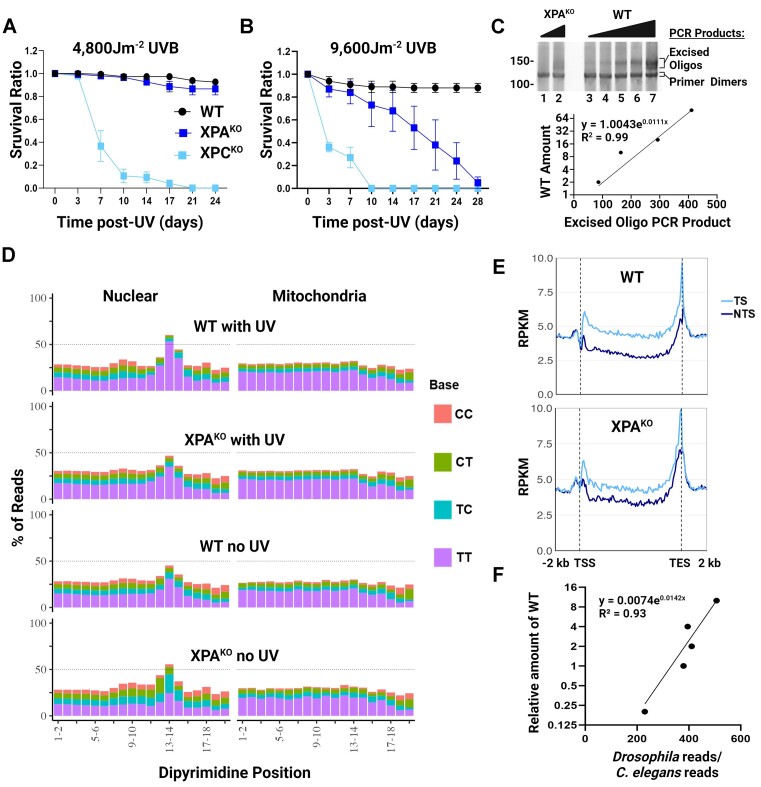
Figure 5.
Clear evidence of excision repair without XPA in Drosophila revealed by CPD qXR-Seq. (A, B) The three indicated fly strains were analyzed for survival for up to four weeks after the indicated doses of UVB. Shown are the mean from three biological replicates with error bars denoting SEM. (C) Analysis of dsDNA libraries of the excised CPD-containing oligos by polyacrylamide gel electrophoresis. Ligation products were PCR-amplified with fifteen cycles, and the PCR product descriptions are indicated on the right, sizes (base pairs) on the left, and quantitation of WT is shown below. (D) Analysis of the frequency of the possible dipyrimidines along reads of 25–30 nt length (trimmed to 20nt from the 5′ end) from the indicated fly strains and UV conditions mapped to either nuclear genome DNA (left) or mitochondrial DNA (right). (E) Analysis of transcription-coupled repair in the WT (top) and XPAKO (bottom) fly strains. CPD qXR-Seq data is plotted as average repair reads (y-axis) along the length of a ‘unit gene’ (x-axis) as described in Figure 3G (The median length for the selected genes is 3024 bp). (F) The spike-in analysis of the fly:worm read ratio from the UV-irradiated WT dilution was used to determine the percentage of excised oligos in the different samples relative to WT. WT, wildtype; KO, knockout; J, Joules; RPKM, reads per kilobase per million mapped reads; TSS, transcription start site; TES, transcription end site; TS, transcribed strand; NTS, nontranscribed strand.