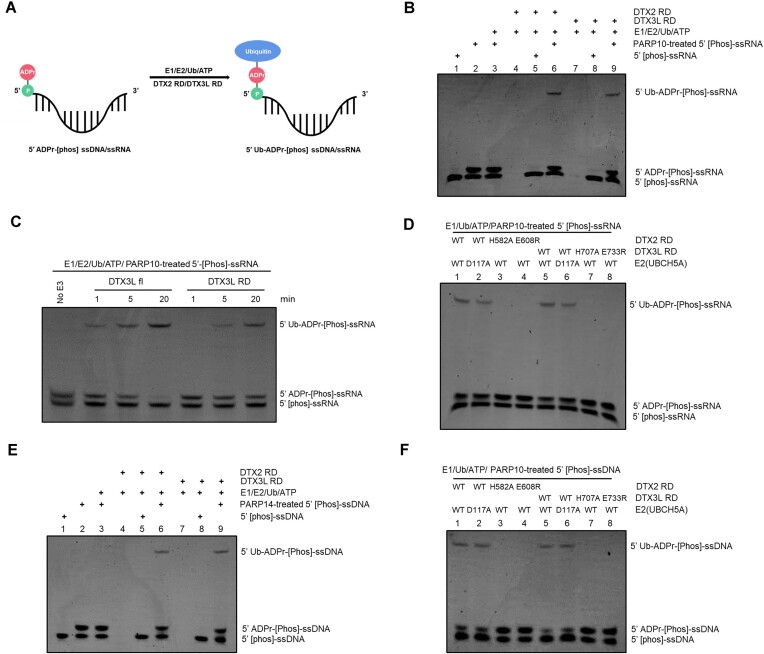
Figure 4.
DTX3L-RD ubiquitylates ADP-ribosylated nucleic acids on the ADPr moiety. (A) A schematic showing the ubiquitylation of ADP-ribosylated nucleic acids. DTX2 RD/DTX3L-RD ubiquitylated 5′ ADP-ribosylated nucleic acids on the ADPr moiety, forming a combined Ub-ADPr- modification. (B) Biochemical reconstitution of the ubiquitylation of ADP-ribosylated ssRNA. ADP-ribosylated and (or) unmodified ssRNA was incubated with either DTX2 RD or DTX3L RD in the presence of Ub processing components (E1, E2, Ub and ATP). The samples were analyzed on a urea gel, which is then visualized under UV light (340 nm) after ethidium bromide (EB) staining. (C) DTX3L full length protein ubiquitylated ADP-ribosylated ssRNA with stronger efficacy than DTX3L RD does. ADP-ribosylated and (or) unmodified ssRNA was incubated with either DTX3L fl or DTX3L RD in the presence of Ub processing components (E1, E2, Ub and ATP). The reactions were stopped at indicated times and analyzed on a urea gel, which is then visualized under UV light (340 nm) after ethidium bromide (EB) staining. (D) DTX2 RD/DTX3L RD ubiquitylated 5′ ADP-ribosylated ssRNA on the ADPr moieties. Asp117 of UbCH5A (E2), which is defective of catalyzing amine group ubiquitylation (68,69), is used to show that DTX2 RD and DTX3L RD ubiquitylated the hydroxyl groups in ADP-ribosylated nucleic acids. DTX2 and DTX3L ADPr ubiquitylation inactive mutants failed to produce upshift bands that correspond to ubiquitylation of ADP-ribosylated nucleic acids, confirming that Ub is attached to ADPr moiety. The samples were analyzed on a urea gel, which is then visualized under UV light (340 nm) after ethidium bromide (EB) staining. (E) As in (B), ssDNA was used as substrate for ubiquitylation assay. (F) As in (D), ubiquitylated ADP-ribosylated ssDNA was used as substrate for hydrolysis assay.