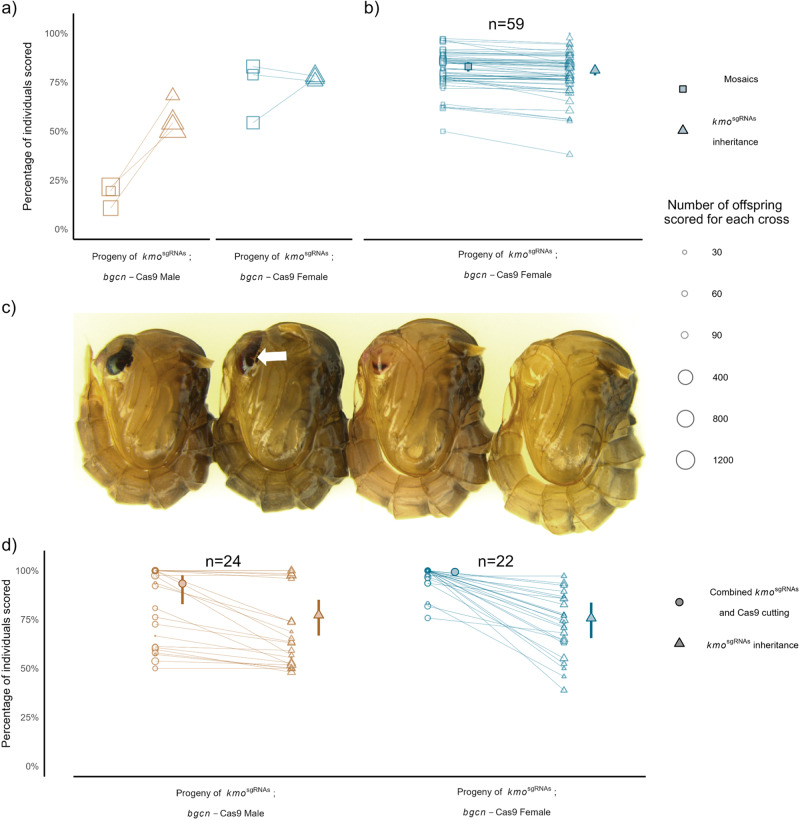
Fig. 2. bgcn-Cas9 biases the inheritance of kmosgRNAs.
a Female bgcn-Cas9 were crossed to kmosgRNAs males (F0) for an initial pooled assessment. Trans-heterozygous F1 males and females were outcrossed to WT in three replicate crosses and the F2 progeny scored for inheritance of the kmosgRNAs transgene (triangles) and eye phenotype (squares). b A replicate cross was completed and the proportion of the F2 progeny inheriting the kmosgRNAs transgene (triangles) and eye phenotype (squares) were scored from individual F1 females. c Images of pupae displaying the different eye phenotypes from left to right: wild type (dark), weak mosaic (white arrow indicating mosaicism), strong mosaic, white eye. d Combined kmosgRNAs inheritance and Cas9 cutting are represented by circle points and are measured as the percentage of offspring with white eyes from crosses between males (n = 24) or females (n = 22) trans-heterozygous for kmosgRNAs; bgcn-Cas9 and kmo−/− mosquitoes. kmosgRNAs inheritance alone is measured as the percentage of offspring with AmCyan fluorescence (triangles). Individual faded points represent offspring from one drive parent, and the size of the point is proportional to the number of offspring from the parent. Horizontal lines connect pools of offspring to show the relationship between cutting and inheritance rates in each replicate. Large symbols and error bars (vertical lines) represent estimated mean and 95% confidence intervals calculated by a generalized linear mixed model, with a binomial (‘logit’ link) error distribution. Data points may be overlapped and some cannot be discerned. Source data are provided as a Source Data file.