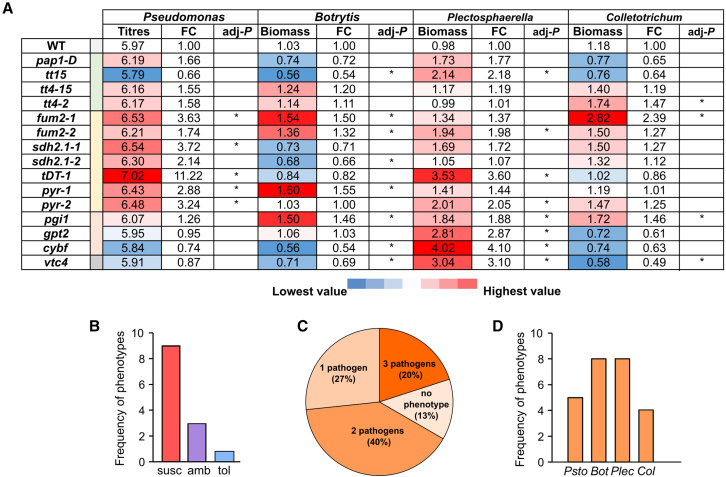
Figure 7.
Overview of immune phenotypes of selected candidates to pathogen infection.
(A) Binary map summarizing the immune phenotypes of the indicated Arabidopsis mutant lines upon challenge with Psto (Pseudomonas), Bot (Botrytis), Plectosphaerella cucumerina (Plectosphaerella), and Colletotrichum higginsianum (Colletotrichum) in Supplemental Figure 10. Color code depicts average colony titer (Psto) or fungal biomass (Botrytis, Plectosphaerella, and Colletotrichum). FC was calculated in comparison to wild-type (WT) plantlets, and asterisks indicate significant differences from the WT (adjusted P 0.05, Duncan’s post hoc test on linear mixed models, n = 4 independent biological experiments).
(B) Frequency of immune phenotypes for selected mutants: enhanced susceptibility (susc), enhanced tolerance (tol), or ambiguous (amb).
(C) Pie chart depicting the fraction of mutants that displayed significant immune phenotypes to one or several pathogens.
(D) The number of mutant alleles that produced immune phenotypes to Pseudomonas (Psto), Botrytis (Bot), Plaectosphaerella (Plec), and Colletotrichum (Col).