Figure 1:
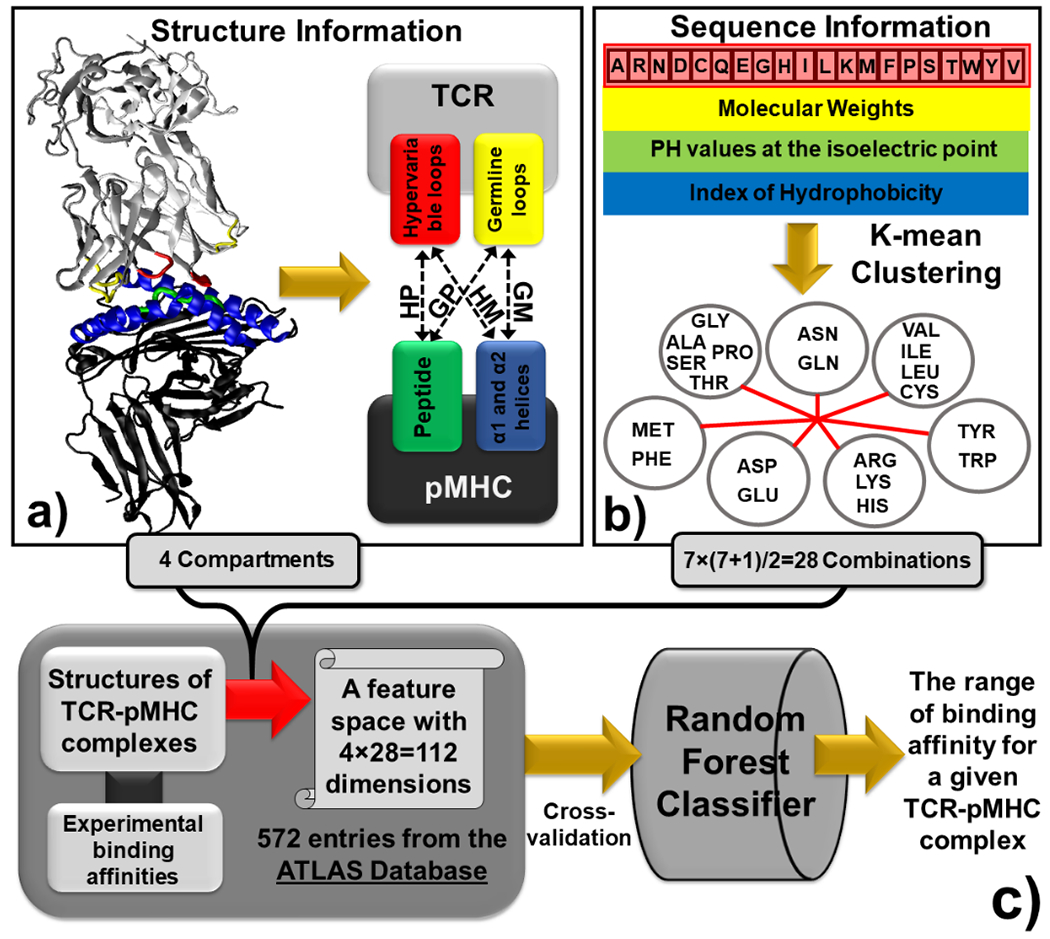
The binding interfaces between TCRs and pMHC are represented by vectors in a high-dimensional feature space. We first divided the structure of a binding interface into four compartments (a). The information of primary sequence at binding interfaces was further integrated into the feature space by coarse-graining the 20 types of amino acids into 7 groups based on the three physicochemical properties (b). Based on the construction of the feature space, all TCR-pMHC complexes in the ATLAS database were used as the benchmark to train a random forest classifier, so that the range of binding affinity for a specific TCR-pMHC complex can be predicted (c).
