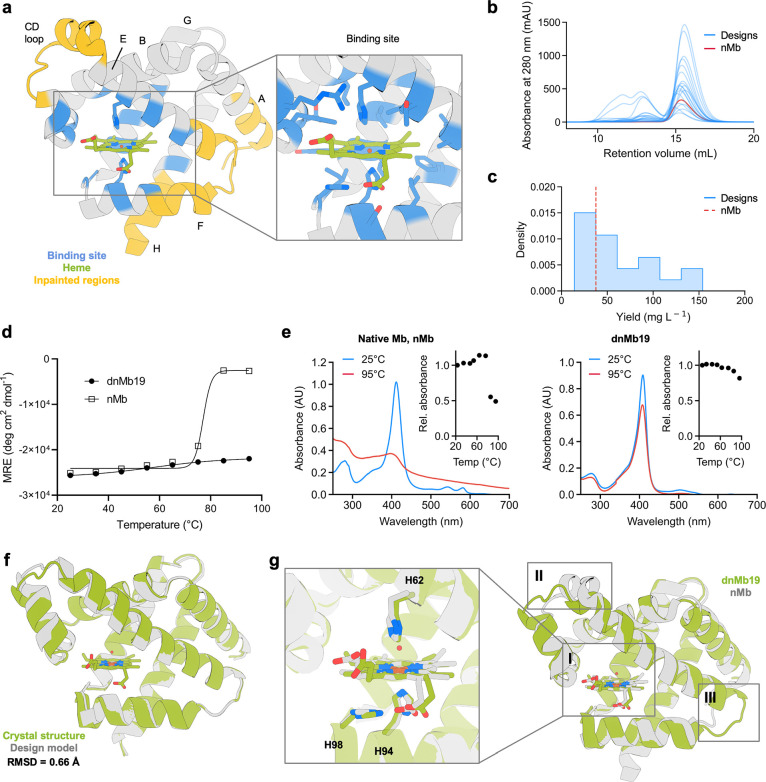
Figure 2.
ProteinMPNN design improves myoglobin expression and thermostability. (a) Positions adjacent to the heme were kept fixed during the sequence design (shown in blue). Non-conserved regions (in yellow) were subjected to backbone remodeling. Inset shows the heme-binding site. (b) SEC traces of 20 designed myoglobin variants. (c) Soluble yield of myoglobin designs and native myoglobin (nMb, represented as a red dashed line). (d) CD melting temperature plots of dnMb19 compared to native myoglobin (signal reported in molar residue ellipticity (MRE)). (e) Absorbance plots of dnMb19 and native myoglobin (inset shows the temperature scan). (f) Structural alignment of the crystal structure (green) and AlphaFold2 (AF2) prediction (gray) of dnMb19. (g) Overlay of the crystal structure of native myoglobin (gray) and the crystal structure of dnMb19 (green, PDB: 8U5A). Non-conserved regions displayed in insets II and III were subjected to backbone redesign.