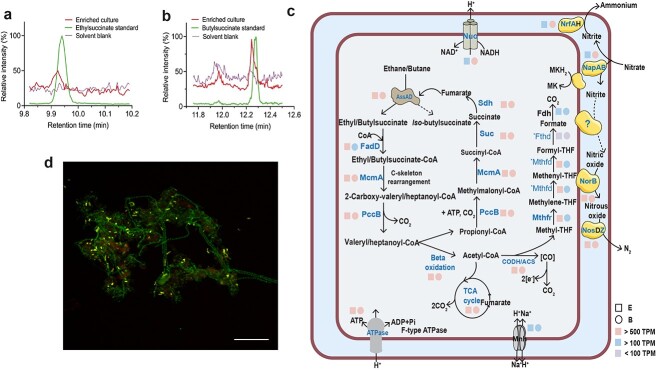
Figure 2.
Metabolic intermediates, inferred metabolic pathways, and a fluorescent micrograph of “Ca. A. nitratireducens”; (A) partial ion chromatograms (ion transition, m/z: 275>73.1) of culture extracts from the C2H6-fed bioreactor displayed a characteristic peak at a retention time of 9.940 min, matching the ethylsuccinate standard; (B) a characteristic peak at a retention time of 12.245 min (ion transition, m/z: 303.0>147.1), consistent with the peak from the butyl-succinate standard, was observed for the culture extracts from the C4H10-fed bioreactor (n = 4 at different sampling points); (C) cell cartoon illustrating “Ca. A. nitratireducens” in the C2H6- or C4H10-fed bioreactors (E or B) use AssA to activate ethane/butane to ethyl/butyl-succinate, which are further converted to acetyl-CoA and propionyl-CoA; fumarate could be regenerated by the methylmalonyl-CoA pathway or the TCA cycle; CO2 is produced through the TCA cycle or the reverse WL pathway; the E and B both harbour genes that enable denitrification (except nirS/K) and dissimilatory nitrate reduction to ammonium; the colour of the square and circle symbols indicates the normalized gene expression values calculated as TPM (total TPM); blue bold text shows that the proteins were fully or partially detected in the protein extracts (*Mthfd and *Fthd were only identified in B and E, respectively), while proteins in black text were not detected; (D) a composite fluorescence micrograph of the C4H10-fed enrichment culture hybridized with the SYMB-1018 probe [16] (Cy3, red; targeting “Ca. A. nitratireducens”) and EUBmix probe set [37] (fluorescein isothiocyanate label, green; all bacteria); “Ca. A. nitratireducens” cells appear yellow (red + green) and other bacterial cells appear green; the scale bar indicates 20 μm; the representative image was selected based on the visual assessment of >3 separate hybridization experiments; FISH was performed as detailed in our previous study [16].