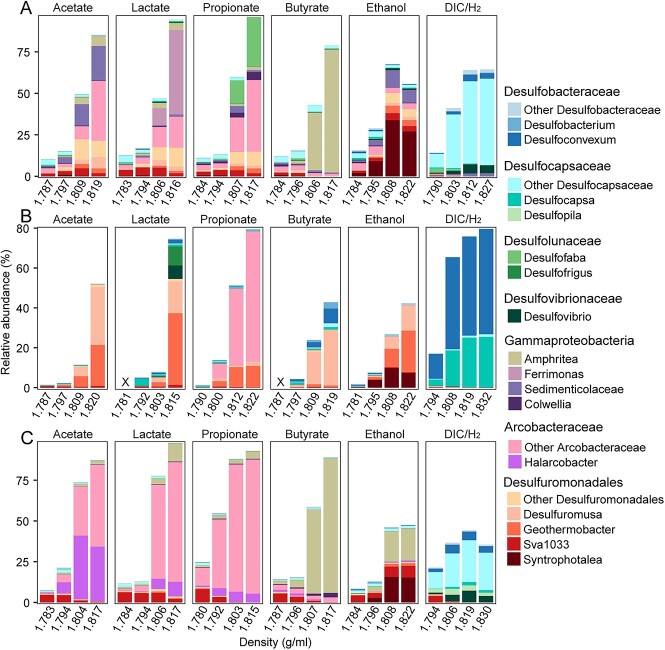
Figure 2.
Identification of the active fermentation products degraders using RNA-SIP in incubations amended with 13C-labeled substrates and sulfate; relative abundance of 16S rRNA gene sequences of active bacteria fermentation product degraders in total bacteria from RNA-SIP gradient fractions in the Helgoland mud (A), Cumberland Bay (B), and Hornsund fjord sediment (C) incubations; × indicates that cDNA synthesis failed because of insufficient amount of RNA in these fractions; density was indicated as the average density of combined fractions for RNA-SIP samples; for sampling time points, see Figure 1.