Abstract
A gram-positive, coryneform bacterium was isolated from swollen scleromata of a dermatosis patient. An analysis of its phenotypic, chemotaxonomic, and genotypic characteristics showed that this bacterium is closely associated with Arthrobacter oxydans and Arthrobacter polychromogenes but that it belongs to a distinct species, for which the name Arthrobacter scleromae sp. nov. is proposed.
CASE REPORT
A 19-year-old male presented to our clinic with a 2-month history of swollen scleromata on his back and hip. He complained of itching and pain at the foci of infection, especially when being touched, and of difficulties sitting and sleeping. No other relevant symptoms were present. While the patient was somewhat thin, his nutritional state was normal. He denied having any other previous illnesses of note or a family history of skin sensitivity.
Scattered, dull red, swollen dermal scleromata on the patient's back and hip were noted upon examination. Some of the scleromata were as large as chicken eggs, had white pus on the tops, and oozed a bloody effusion when pierced. No lymphadenopathy was detected, and the physical examination was otherwise normal. Routine hematological investigations revealed a leukocyte count of 6.5 × 109 cells/liter, with 38.5% lymphocytes, a hemoglobin level of 12.6 g/dl, and a platelet count of 250 × 109/liter. The lymphocyte T4/T8 ratio was 1.5. The serum immunoglobulin G (IgG) level was 1,120 mg/dl, the IgA level was 410 mg/dl, and the IgM level was 183 g/dl. Complement C3 was at 0.9 g/liter, C4 was at 0.75 g/liter, and CH50 was at 66.6 U. The serum HBsAg test was negative. The scleromata developed slowly and looked like granuloma. No skin biopsy was performed. Samples of the bloody effusion were sent for bacterial, mycobacterial, and fungal cultures. A rapidly growing bacterium with numerous colonies in a pure culture state on blood agar was seen after 24 h at 35°C. The isolate was suspected of being Staphylococcus epidermidis due to colonial and cell morphology. But in succedent liquid culture, a rod coccus growth cycle was observed within 24 h at 35°C. Thus, a gram-positive coryneform bacterium, which was subsequently identified as an Arthrobacter sp., was recognized. The patient was treated with oral Chinese herbal medicine for heat clearing and detoxifying once a week for 1 month. Meanwhile, empirical antimicrobial therapy with oral ampicillin was initiated. Once the culture result was ascertained, on the fifth day from the start of the therapy, antibiotic treatment was changed to intravenous ceftriaxone and cefazolin for 2 weeks and then to oral ceftriaxone, with slow resolution of the lesion over a 2-month period.
The genus Arthrobacter includes a heterogeneous group of aerobic, gram-positive, catalase-positive, nonfermentative coryneform bacteria of high G+C DNA content (18). Members of this genus contain l-lysine as the diamino acid in their cell walls and major cellular fatty acids (CFAs) of the branched type (11) and are divided into two groups, the A3α and A4α variations, based on their peptidoglycan structure (28). Arthrobacter spp. are widely distributed in the environment, especially in soil, and have recently been recognized as opportunistic pathogens. Among the 38 validly described Arthrobacter species at the time of writing of the present report, 5 were isolated from clinical sources, namely Arthrobacter albus (30), Arthrobacter creatinolyticus (13), Arthrobacter cumminsii (10), Arthrobacter luteolus (30), and Arthrobacter woluwensis (10). In addition, some strains of Arthrobacter oxydans were isolated from human blood (30). However, knowledge of the distribution, pathogenic potential, and clinical significance of Arthrobacter strains is far from adequate. The discovery of a new Arthrobacter species of human origin is therefore still significant. In this investigation, we report the characteristics of an unknown Arthrobacter species isolated from clinical specimens. On the basis of phenotypic, chemotaxonomic, and genotypic data, we propose the description of a new species of the genus Arthrobacter, Arthrobacter scleromae.
Strain YH-2001T (AS 1.3601T; China General Microbiological Culture Collection Center, Chinese Academy of Sciences) was isolated from the bloody effusion from swollen dermal scleromata of the patient. Additionally, two reference strains, Arthrobacter oxydans AS 1.1925T (DSM 20119T) and Arthrobacter polychromogenes AS 1.1927T (DSM 20136T), were included for comparison in this study. These organisms were inoculated on nutrient agar and incubated at 30°C.
The morphology and motility of cells were examined by using a model FEI QUANTA electron microscope and a Zeiss Axioskop 20 light microscope. Motility was tested by the hanging-drop technique (29). Flagellum staining was performed as described by Kodaka et al. (19). Biochemical tests, including utilization of carbohydrates as sole carbon sources, were performed by using either conventional methods (11, 12) or commercially available API Coryne and API 50 CH kits (bioMérieux, Marcy l'Étoile, France) according to the manufacturer's instructions.
Antibiotic susceptibility was determined by the E test on Mueller-Hinton blood agar incubated at 35°C for 24 h. The results were interpreted according to the criteria established for staphylococci in 1997 by the National Committee for Clinical Laboratory Standards (24).
Cell-wall diamino acids and sugars were prepared and analyzed as described by Komagata and Suzuki (21). Detailed analysis of the peptidoglycan structures was performed by following the methods of Schleifer and Kandler (28). The amino acids and peptides were separated by two-dimensional ascending thin-layer chromatography on cellulose sheets (Merck) by using the solvent systems described by Schleifer and Kandler (28). Menaquinones were extracted from freeze-dried biomass, purified according to the method of Collins (4), and then analyzed by high-pressure liquid chromatography. For analysis of CFAs, cells of the test strain were cultured on trypticase soy agar (BBL) for 48 h at 30°C. CFAs were extracted, methylated, and analyzed by gas chromatography by using the standard Microbial Identification System (15). The base composition of the genomic DNA of the strain was determined by the thermal denaturation method (23) with Escherichia coli AS 1.365 as a control.
Genomic DNA preparation and PCR amplification of the 16S rRNA gene were performed by using the method of Chun and Goodfellow (3). The amplified product was sequenced as described previously (14). The nucleotide sequence was obtained automatically by using an Applied Biosystems DNA sequencer (model 377) and software provided by the manufacturer. The 16S rRNA gene sequence of strain YH-2001T was aligned manually by using the CLUSTAL X 1.8 program against corresponding sequences retrieved from the GenBank database. Phylogenetic trees were inferred by using the neighbor-joining (26), least-squares (9), and maximum-likelihood (6) treeing algorithms from the PHYLIP 3.5c suite of programs (8). Evolutionary distance matrices were generated by the method of Kimura (16). The resultant unrooted tree topologies were evaluated by bootstrap analyses (7) of the neighbor-joining method based on 1,000 resamplings with the SEQBOOT and CONSENSE programs in the PHYLIP package.
Hybridizations between genomic DNA of strain YH-2001T and that of its nearest neighbors were carried out by following the micro-well method (2, 5) with a FLUOstar OPTIMA microplate reader (BMG LABTECH, Offenburg, Germany) for the fluorescence measurements. Single-stranded unlabeled DNA was noncovalently bound to microplate wells. Hybridizations were performed at 47°C overnight in a hybridization solution containing 50% formamide but no dextran sulfate. Salmon sperm DNA was used as the negative control in all experiments. Each experiment was performed with four replicates. The percentage of DNA homology was calculated as described by Christensen et al. (2). DNA-DNA homology data generated in this study are presented as means of reciprocal values. The DNA homology value in individual experiments never showed more than 10% deviation from the mean.
Strain YH-2001T consisted of gram-positive, non-spore-forming, nonmotile, coryneform cells, which displayed a typical rod-coccus life cycle. It was aerobic, catalase-positive, and nonfermentative, producing white, glistening, circular colonies with a smooth convex surface. The colonies were able to grow to up to 5.0 mm in diameter after a 72-h incubation. The strain contained l-lysine as the diamino acid in its cell wall and a predominant amount of anteiso-C15:0 as the major CFA, but it lacked mycolic acids. These general properties were consistent with members of the genus Arthrobacter. Results from API test systems showed that the organism was positive for gelatin liquefaction, casein, DNA, esculin, starch, and tyrosine hydrolysis; weakly positive for nitrate reduction; and negative for N-acetylglucosaminidase, lecithinase, lipase, and urease. No acid was produced from the carbohydrates tested, except mannitol when grown aerobically. The organism assimilated l-arginine, l-asparagine, citrate, d-fructose, d-galactose, d-gluconate, d-glucose, glycerol, l-histidine, lactose, maltose, mannose, d-melezitose, pyruvate, d-raffinose, sorbitol, sucrose, trehalose, xylitol, and d-xylose.
The isolate possessed a peptidoglycan of type A3α with an l-Lys-l-Ser-l-Thr-l-Ala interpeptide bridge, which is found in four members of the genus Arthrobacter, i.e., A. chlorophenolicus, A. oxydans, A. polychromogenes, and A. sulfonivorans (1, 20, 32). However, strain YH-2001T had a markedly different isoprenoid quinone profile, composed of a major amount of MK-8(H2) and a minor amount of MK-10 (peak area ratio, 88:12). It was also easily distinguished from A. chlorophenolicus and A. sulfonivorans by colony color, motility, and growth ability at 5°C. A comparison between the chemotaxonomic and phenotypic characteristics of strain YH-2001T and those of the four Arthrobacter species mentioned above is shown in Table 1.
TABLE 1.
Comparison of characteristics of Arthrobacter species with Lys-Ser-Thr-Ala-type peptidoglycana
| Characteristic | Result for organism indicated
|
||||
|---|---|---|---|---|---|
| A. scleromae | A. chlorophenolicus | A. oxydans | A. polychromogenes | A. sulfonivorans | |
| Cell-wall sugars | Gal, Glu | ND | Gal, Glu | Gal | ND |
| Major menaquinone | MK-8(H2) | ND | MK-9(H2) | MK-9(H2) | MK-9(H2) |
| CFA composition (%): | |||||
| i-C14:0 | 1 | 4 | 2 | 1 | 1 |
| C14:0 | 1 | 2 | 2 | 1 | 1 |
| i-C15:0 | 11 | 7 | 9 | 3 | 15 |
| ai-C15:0 | 50 | 66 | 45 | 44 | 63 |
| C15:0 | − | − | 1 | 1 | − |
| i-C16:1 | 4 | 1 | − | − | − |
| i-C16:0 | 7 | 10 | 11 | 6 | 6 |
| C16:1ω7c | 5 | 1 | − | − | − |
| C16:0 | 2 | 3 | 11 | 13 | 4 |
| i-C17:1ω9c | 2 | − | − | − | − |
| ai-C17:1ω9c | 7 | 1 | − | − | − |
| i-C17:0 | 1 | − | 2 | 1 | 1 |
| ai-C17:0 | 8 | 5 | 17 | 25 | 7 |
| G+C content of DNA (mol%) | 64.7 | 65.1 | 63.1 | 62.9 | 61 |
| Colony color | White | Grey | Grey-white | Blue-green | Creamy yellow |
| Motility | − | + | − | − | + |
| Growth at: | |||||
| 5°C | − | + | − | − | + |
| 37°C | + | + | + | + | − |
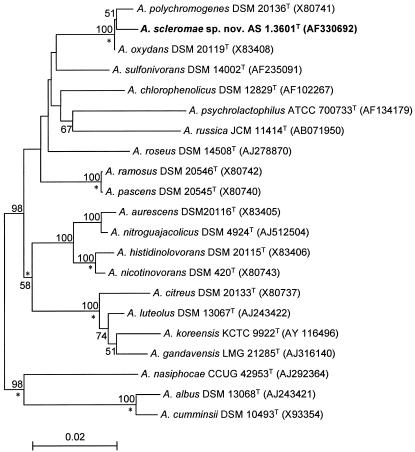
To further clarify the relationships between this unknown bacterium and the recognized Arthrobacter species, its almost complete 16S rRNA gene sequence (1,415 nucleotides) was determined and compared with those of representative reference strains of the genus Arthrobacter. The result clearly showed that the organism belongs to this genus. In accordance with the result of cell-wall peptidoglycan analysis, the bacterium formed a closely associated clade with A. oxydans and A. polychromogenes, supported by a high bootstrap value of 100% (Fig. 1). High 16S rRNA gene sequence similarities of 99.4 (A. oxydans DSM 20119T) and 99.3% (A. polychromogenes DSM 20136T) were found between the test strain and its two nearest neighbors. But this result did not indicate that they belong to the same genospecies, as higher 16S rRNA gene sequence similarities were recorded between representatives of several well-established Arthrobacter species, for example, 99.7% between Arthrobacter nitroguajacolicus and Arthrobacter aurescens (22) and 100% between Arthrobacter pascens and Arthrobacter ramosus (17). Actually, strain YH-2001T exhibited a number of characteristics distinct from its nearest phylogenetic relatives (Table 2). Despite the fact that some strains of A. oxydans were also isolated from human clinical specimens (30), strain YH-2001T differed from A. oxydans by lacking β-galactosidase, α-glucosidase, and the capability to grow on nicotine and on 10% NaCl. The API Coryne code for the strain was 1042014, which was identified as a doubtful or unacceptable profile for Rhodococcus spp. in the manufacturer's database, whereas the API Coryne code for A. oxydans was 3750004 (Arthrobacter spp.) (30). A. polychromogenes was isolated from air, and it forms blue-pigmented colonies on carbohydrate-peptone medium (27), a feature distinguishing it from the other Arthrobacter species and from strain YH-2001T as well. Furthermore, unlike A. oxydans and A. polychromogenes, the test strain could not utilize 2,3-butanediol, formate, l-leucine, malonate, l-rhamnose, or d-ribose as the sole carbon source, nor could it produce acid from glucose, but it was able to grow in mineral salt medium with ammonium or nitrate as the sole nitrogen source, while A. oxydans and A. polychromogenes required biotin (32).
FIG. 1.
Unrooted neighbor-joining tree showing the phylogenetic position of A. scleromae sp. nov. AS 1.3601T within the genus Arthrobacter. Asterisks indicate branches that were also recovered by using the least-squares and maximum-likelihood methods. The numbers at the nodes indicate the levels of bootstrap support (%) based on a neighbor-joining analysis of 1,000 resampled data sets; only the values over 50% are given. The scale bar indicates 0.02 substitutions per nucleotide position.
TABLE 2.
Biochemical characteristics distinguish Arthrobacter scleromae from its nearest phylogenetic relativesa
| Species | Results with:
|
Utilization of:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nic | NO3 | 10% NaCl | Sta | Vit | Glu | β-Gal | α-Glu | l-Leu | l-Ara | l-Rha | d-Rib | 2,3-But | Ino | Mal | For | Pro | |
| A. oxydans | + | + | + | + | + | + | + | + | W | + | + | + | W | W | + | + | + |
| A. polychromogenes | − | + | − | W | + | + | ND | ND | W | + | + | + | W | − | + | + | + |
| A. scleromae | − | W | − | + | − | − | − | − | − | W | − | − | − | W | − | − | W |
Data were taken from this study, Kodama et al. (20), and Wauters et al. (30). +, positive or present; W, weak positive; −, negative or absent; ND, not determined; Nic, utilization of nicotine; NO3, nitrate reduction; 10% NaCl, growth on 10% (wt/vol) NaCl; Sta, hydrolysis of starch; Vit, vitamin requirement; Glu, acid from glucose; β-Gal, β-galactosidase; α-Glu, α-glucosidase; l-Leu, l-leucine; l-Ara, l-arabinose; l-Rha, l-rhamnose; d-Rib, d-ribose; 2,3-But, 2,3-butanediol; Ino, inositol; Mal, malonate; For, formate; Pro, propionate.
DNA-DNA reassociation values between strain YH-2001T and A. oxydans DSM 20119T and A. polychromogenes DSM 20136T were 51 and 36%, respectively. These values are well below the 70% threshold for delineation of genomic species (31), thereby underpinning the idea that the unknown isolate should be assigned to a separate species. On the basis of phenotypic, chemotaxonomic, phylogenetic, and DNA-DNA hybridization evidence, strain YH-2001T merits a new species status in the genus Arthrobacter, for which we propose the name Arthrobacter scleromae sp. nov. Since it was isolated from several cultures of bloody effusion from dermal scleromata of a patient and numerous colonies in a pure state were present at primary isolation, it was not considered to be an environmental contaminant. We believe that the formal description of this new Arthrobacter species will facilitate its recognition in the clinical laboratory and contribute to a better understanding of the role of Arthrobacter strains in human disease.
Description of Arthrobacter scleromae sp. nov.
Arthrobacter scleromae (scle′ro.mae. L. adj. scleromae, of scleroma) cells are gram-positive, non-spore-forming, and nonmotile and display a rod-coccus life cycle. They are obligately aerobic, catalase-positive, and ∼0.25 to ∼0.35 μm in diameter. Colonies on blood agar or nutrient agar are whitish, glistening, convex, smooth surfaced, and circular. The colonies grow to up to 4 to 5 mm in size by 72 h. Growth occurs with a suitable carbon source in mineral salt medium; no additional growth factors are needed. Growth also occurs in the presence of 5% NaCl, at between 15 and 37°C and between pHs 6 and 9, but not on 10% NaCl, at 5 or 42°C. The organism hydrolyzes casein, DNA, esculin, gelatin, starch, and tyrosine, but not lecithin or xanthine. The nitrate reduction test is weakly positive. N-acetylglucosaminidase, β-galactosidase, α-glucosidase, lipase, pyrrolidonyl peptidase, and urease are not produced. Acid is produced from mannitol. It utilizes the following substrates as sole carbon sources: γ-aminobutyrate, l-arginine, l-asparagine, citrate, d-fructose, fumarate, d-galactose, d-gluconate, d-glucose, glycerol, l-histidine, p-hydroxybenzoate, 2-ketoglutarate, lactose, dl-malate, maltose, mannose, d-melezitose, phenylacetate, pyruvate, d-raffinose, salicin, sorbitol, succinate, sucrose, trehalose, d-turanose, xylitol, and d-xylose. The following substrates are not utilized: acetamide, adipate, adonitol, l-alanine, p-aminobenzoate, d-arabitol, arbutin, azelate, benzoate, 2,3-butanediol, n-butyrate, caprylate, d-cellobiose, l-citrulline, l-cysteine, dulcitol, erythritol, formate, glucosamine, d-glucuronate, glutarate, glycogen, glycolate, γ-hydroxybutyrate, inulin, dl-isoleucine, isovalerate, l-leucine, maleate, malonate, dl-methionine, α-methyl-d-glucoside, α-methyl-d-mannoside, nicotine, l-ornithine, oxalate, l-phenylalanine, o-phthalate, pimelate, l-rhamnose, d-ribose, sebacate, sorbose, suberate, d-tartrate, l-threonine, l-tryptophan, or dl-valine. Compounds slowly utilized are acetate, l-arabinose, inositol, mannitol, and melibiose. The strain is susceptible to ceftriaxone, chloramphenicol, rifampin, and tetracycline; moderately susceptible to cefazolin, cefotaxime, doxycycline, erythromycin, nitrofurantoin, piperacillin, and vancomycin; and resistant to amikacin, ampicillin, gentamicin, kanamycin, norfloxacin, oxacillin, penicillin G, streptomycin, and tobramycin. The DNA G+C content is 64.7 mol%. The major CFA is anteiso-C15:0, with significant amounts of i-C15:0, i-C16:0, ai-C17:1ω9c, and ai-C17:0. The predominant menaquinone is MK-8(H2). The cell-wall peptidoglycan type is l-Lys-l-Ser-l-Thr-l-Ala (A3α), and the cell-wall sugars are galactose and glucose. It was isolated from swollen scleromata of a dermatosis patient. The type strain is AS 1.3601T.
Nucleotide sequence accession number.
The GenBank accession number for the 16S rRNA gene sequence of Arthrobacter scleromae AS 1.3601T is AF330692.
REFERENCES
- 1.Borodina, E., D. P. Kelly, P. Schumann, F. A. Rainey, N. L. Ward-Rainey, and A. P. Wood. 2002. Enzymes of dimethylsulfone metabolism and the phylogenetic characterization of the facultative methylotrophs Arthrobacter sulfonivorans sp. nov., Arthrobacter methylotrophus sp. nov., and Hyphomicrobium sulfonivorans sp. nov. Arch. Microbiol. 177:173-183. [DOI] [PubMed] [Google Scholar]
- 2.Christensen, H., O. Angen, R. Mutters, J. E. Olsen, and M. Bisgaard. 2000. DNA-DNA hybridization determined in micro-wells using covalent attachment of DNA. Int. J. Syst. Evol. Microbiol. 50:1095-1102. [DOI] [PubMed] [Google Scholar]
- 3.Chun, J., and M. Goodfellow. 1995. A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 45:240-245. [DOI] [PubMed] [Google Scholar]
- 4.Collins, M. D. 1985. Isoprenoid quinone analysis in classification and identification, p. 267-287. In M. Goodfellow and D. E. Minnikin (ed.), Chemical methods in bacterial systematics. Academic Press, London, United Kingdom.
- 5.Ezaki, T., Y. Hashimoto, and E. Yabuuchi. 1989. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39:224-229. [Google Scholar]
- 6.Felsenstein, J. 1981. Evolutionary tree form DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1993. PHYLIP (phylogenetic inference package) version 3.5c. Department of Genetics, University of Washington, Seattle, Wash.
- 9.Fitch, W. M., and E. Margoliash. 1967. Construction of phylogenetic trees: a method based on mutation distances as estimated from cytochrome c sequences is of general applicability. Science 155:279-284. [DOI] [PubMed] [Google Scholar]
- 10.Funke, G., R. A. Hutson, K. A. Bernard, G. E. Pfyffer, G. Wauters, and M. D. Collins. 1996. Isolation of Arthrobacter spp. from clinical specimens and description of Arthrobacter cumminsii sp. nov. and Arthrobacter woluwensis sp. nov. J. Clin. Microbiol. 34:2356-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Funke, G., A. von Graevenitz, J. E. Clarridge III, and K. A. Bernard. 1997. Clinical microbiology of coryneform bacteria. Clin. Microbiol. Rev. 10:125-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holding, A. J., and J. G. Collee. 1971. Routine biochemical tests. Methods Microbiol. 6A:2-32. [Google Scholar]
- 13.Hou, X. G., Y. Kawamura, F. Sultana, S. Shu, K. Hirose, K. Goto, and T. Ezaki. 1998. Description of Arthrobacter creatinolyticus sp. nov., isolated from human urine. Int. J. Syst. Bacteriol. 48:423-429. [DOI] [PubMed] [Google Scholar]
- 14.Huang, Y., W. Qi, Z. Lu, Z. Liu, and M. Goodfellow. 2001. Amycolatopsis rubida sp. nov., a new Amycolatopsis species from soil. Int. J. Syst. Evol. Microbiol. 51:1093-1097. [DOI] [PubMed] [Google Scholar]
- 15.Kämpfer, P., and R. M. Kroppenstedt. 1996. Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can. J. Microbiol. 42:989-1005. [Google Scholar]
- 16.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 17.Koch, C., F. A. Rainey, and E. Stackebrandt. 1994. 16S rDNA studies on members of Arthrobacter and Micrococcus: an aid for their taxonomic restructuring. FEMS Microbiol. Lett. 123:167-172. [Google Scholar]
- 18.Koch, C., P. Schumann, and E. Stackebrandt. 1995. Reclassification of Micrococcus agilis (Ali-Cohen 1889) to the genus Arthrobacter as Arthrobacter agilis comb. nov. and emendation of the genus Arthrobacter. Int. J. Syst. Bacteriol. 45:837-839. [DOI] [PubMed] [Google Scholar]
- 19.Kodaka, H., A. Y. Armfield, G. L. Lombard, and V. R. Dowell, Jr. 1982. Practical procedure for demonstrating bacterial flagella. J. Clin. Microbiol. 16:948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kodama, Y., H. Yamamoto, N. Amano, and T. Amachi. 1992. Reclassification of two strains of Arthrobacter oxydans and proposal of Arthrobacter nicotinovorans sp. nov. Int. J. Syst. Bacteriol. 42:234-239. [DOI] [PubMed] [Google Scholar]
- 21.Komagata, K., and K.-I. Suzuki. 1987. Lipid and cell-wall analysis in bacterial systematics, p. 161-207. In R. R. Colwell and R. Grigorova (ed.), Methods in microbiology, vol. 19. Academic Press, New York, N.Y. [Google Scholar]
- 22.Kotouèková, L., P. Schumann, E. Durnová, C. Spröer, I. Sedláèek, J. Neèa, Z. Zdráhal, and M. Nìmec. 2004. Arthrobacter nitroguajacolicus sp. nov., a novel 4-nitroguaiacol-degrading actinobacterium. Int. J. Syst. Evol. Microbiol. 54:773-777. [DOI] [PubMed] [Google Scholar]
- 23.Marmur, J., and P. Doty. 1962. Determination of base composition of deoxyribonucleic acid from its denaturation temperature. J. Mol. Biol. 5:109-118. [DOI] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 1997. Minimum inhibitory concentration (MIC) interpretive standards (μg/ml) for organisms other than Haemophilus spp., Neisseria gonorrhoeae, and Streptococcus spp. NCCLS document M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 25.Reddy, G. S., J. S. Prakash, G. I. Matsumoto, E. Stackebrandt, and S. Shivaji. 2002. Arthrobacter roseus sp. nov., a psychrophilic bacterium isolated from an Antarctic cyanobacterial mat sample. Int. J. Syst. Evol. Microbiol. 52:1017-1021. [DOI] [PubMed] [Google Scholar]
- 26.Saitou, N., and M. Nei. 1987. The neighbor joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 27.Schippers-Lammertse, A. F., A. O. Muijsers, and K. B. Klatser-Oedekerk. 1963. Arthrobacter polychromogenes nov. spec., its pigments, and a bacteriophage of this species. Antonie Leeuwenhoek 29:1-15. [DOI] [PubMed] [Google Scholar]
- 28.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skerman, V. B. K. 1967. A guide to the identification of the genera of bacteria, 2nd ed. Williams & Wilkins, Baltimore, Md.
- 30.Wauters. G., J. Charlier, M. Janssens, and M. Delmee. 2000. Identification of Arthrobacter oxydans, Arthrobacter luteolus sp. nov., and Arthrobacter albus sp. nov., isolated from human clinical specimens. J. Clin. Microbiol. 38:2412-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackenbrandt, M. P. Starr, and H. G. Trüper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 32.Westerberg, K., A. M. Elvang, E. Stackebrandt, and J. K. Jansson. 2000. Arthrobacter chlorophenolicus sp. nov., a new species capable of degrading high concentrations of 4-chlorophenol. Int. J. Syst. Evol. Microbiol. 50:2083-2092. [DOI] [PubMed] [Google Scholar]