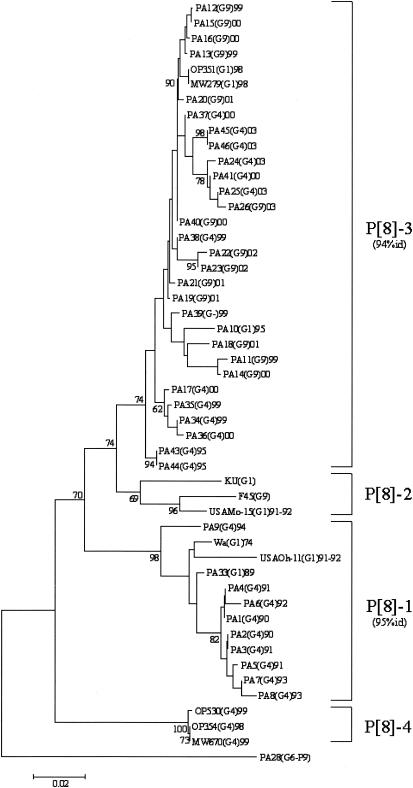
FIG. 3.
Phylogenetic analysis of partial VP4 nucleotide sequences (nt 29 to 449) of type P[8] strains. The phylogenetic tree was constructed using the neighbor-joining method and Kimura two-parameter model. Type P [9] PA28 sequence was used as an outgroup. Percentage bootstrap values above 60% are given at branch nodes. Nucleotide identity (id) within lineages is indicated in parentheses to the right of the lineage. The number of substitutions per site is indicated by the scale bar.