Abstract
Borrelia lusitaniae is a species within the complex Borrelia burgdorferi sensu lato and is infrequently isolated in Europe. In contrast, this species is by far the most predominant in North Africa and in Portugal. In this study, we analyzed the genetic diversity, at several loci, of a large population of isolates from free-living Ixodes ricinus ticks collected in Tunisia and Morocco. We found a moderate diversity of the whole genome by using pulsed-field gel electrophoresis as well as in the ospA gene sequences, compared to a high level of strain homogeneity in the small noncoding ribosomal spacer. In contrast, a high diversity of this locus has been previously reported for Portuguese isolates. We hypothesize that B. lusitaniae strains isolated in North Africa constitute a clone of Portuguese origin.
Within the complex Borrelia burgdorferi sensu lato, three species, B. burgdorferi sensu stricto, Borrelia garinii, and Borrelia afzelii, are known to cause a broad spectrum of human manifestations that are described as Lyme borreliosis. The genetic diversity of pathogenic species has been largely investigated both at the species level and at the strain level within each species (41). Otherwise, although its pathogenicity for humans is only suspected (31, 33), Borrelia valaisiana represents a widely distributed species in ticks throughout Europe and Asia and therefore has been extensively studied (12, 22, 32, 38, 40).
In contrast to these species, little data are available concerning Borrelia lusitaniae. Only sporadic isolates have been characterized from ticks collected in the Czech Republic, Moldavia, Ukraine, and Belarus (20), in Slovakia (11), in Spain (3), in Poland (25), in France (30), in Switzerland (15-17), or in Turkey (13), and these isolates did not show any significant diversity. However, a study conducted in Portugal revealed a large diversity among B. lusitaniae strains isolated from a local tick population (6). Moreover, it is noteworthy that, among the three first Portuguese isolates identified as B. lusitaniae, two different types were demonstrated (20, 26). Hence, these studies suggested a higher diversity among B. lusitaniae around the Mediterranean basin. However, all these studies only focused on the small ribosomal rrf-rrl spacer and, therefore, the global diversity of this species remained unknown.
Recently, we reported that B. lusitaniae was, by far, the most prevalent species, ranging from 96.6 to 100% of the B. burgdorferi sensu lato species identified in Tunisia and Morocco (35, 44, 46). The report of some Lyme borreliosis cases in North Africa (1, 27) and the recent isolation in Portugal of B. lusitaniae from the skin of a human patient with chronic lesions (5) prompted us to characterize this species as a potential human pathogen. Therefore, we investigated the molecular characterization and the genetic diversity of B. lusitaniae isolated from free-living Ixodes ricinus ticks in Tunisia and Morocco. For these purposes we studied two single loci (ospC and ospA) and the noncoding intergenic spacer (rrf-rrl), as well as the whole genome.
MATERIALS AND METHODS
Study area and tick collection.
The study was carried out in two sites in Tunisia (44). Jbel el Jouza is located in a lower-humidity zone with mild winters where the rainfall ranges from 600 to 800 mm/year in Northwest, and Ain Draham is in a higher-humidity bioclimatic zone in Northwest where the rainfall is >800 mm/year. In Morocco, the study was conducted in Taza, a humid area in the middle Occidental atlas, which was the only site that revealed the presence of I. ricinus ticks (35).
Samplings at these sites were conducted between 2000 and 2003. Questing ticks were collected by blanket dragging the vegetation, as previously reported (35, 44). Only adult ticks were used in this study.
Borrelia culture and DNA extraction.
Live field-collected I. ricinus ticks were dipped in 70% ethanol, rinsed in distilled water, and then ground up and inoculated into 2 ml of BSK-H medium (Sigma) supplemented with 7% gelatin and 1% antibiotic mixture for Borrelia (Sigma). Tightly capped tubes were incubated at 34°C and examined once a week by dark-field microscopy. Subcultures were performed without antibiotics and into a larger volume. For pulsed-field gel electrophoresis (PFGE) only isolates maintained in pure cultures were used, whereas for PCR experiments contaminated cultures and/or cultures containing few spirochetes were used to extract DNA by boiling at 100°C for 10 min. Strains and DNA used in this study are shown in Table 1.
TABLE 1.
B. lusitaniae from Maghrebin countries used in this work and the targets studied for each isolatea
| Isolate | rrf-rrl spacerb | PFGE | ospC | ospAb |
|---|---|---|---|---|
| Tunisian isolates | ||||
| TD 12 | AY575748 | X | X | |
| TT 4 | AY575747 | X | X | |
| TT 13 | AY575749 | X | ||
| TT 214 | AY575755 | X | ||
| TT 266 | AY575756 | X | ||
| TT 328 | X | |||
| TT 39 | AY575750 | X | ||
| TT 40 | AY575751 | X | ||
| TT 840 | AY575758 | X | X | AJ634238 |
| TT 85 | AY575754 | X | ||
| TT 867 | AY575759 | X | X | AJ634239 |
| TT 892 | X | |||
| TT 894 | AY575760 | X | X | |
| TT 899 | X | X | ||
| TT 908 | AY575761 | X | X | AJ634240 |
| TT 916 | AY575762 | X | X | AJ634241 |
| TT 917 | X | |||
| TT 918 | AY575763 | X | X | |
| TT 919 | X | X | ||
| TT 920 | AY575764 | X | X | |
| TT 925 | AY575765 | X | AJ634242 | |
| TT 927 | AY575766 | X | ||
| TT 928 | AY575767 | X | AJ634243 | |
| TT57 | AY575753 | |||
| TT55 | AY575752 | |||
| TT832 | AY575757 | |||
| Moroccan isolates | ||||
| MT1 | AY575720 | X | ||
| MT2 | AY575721 | X | AJ634228 | |
| MT4 | AY575722 | X | X | AJ634229 |
| MT6 | AY575723 | |||
| MT7 | AY575724 | X | AJ634230 | |
| MT8 | AY575725 | X | X | AJ634231 |
| MT9 | X | X | AJ634232 | |
| MT10 | AY575726 | X | X | AJ634233 |
| MT11 | AY575727 | X | X | |
| MT12 | AY575728 | X | X | AJ634234 |
| MT13 | X | X | ||
| MT14 | AY575729 | X | ||
| MT16 | AY575730 | X | X | AJ634235 |
| MT17 | AY575731 | X | ||
| MT18 | AY575732 | X | ||
| MT20 | AY575733 | X | X | |
| MT22 | AY575734 | X | X | |
| MT23 | AJ634236 | |||
| MT24 | AY575735 | X | X | |
| MT25 | AY575736 | X | ||
| MT26 | AY575737 | X | AJ634237 | |
| MT27 | AY575738 | X | X | |
| MN2-6 | AY575741 | |||
| MN2-29 | AY575744 | |||
| MN2-32 | AY575740 | |||
| M3-3 | AY575745 | |||
| M3-4 | AY575746 | |||
| M3-22 | AY575742 | |||
| MA3-13 | AY575739 | |||
| MA3-45 | AY575743 |
Only isolates maintained in pure culture were analyzed by PFGE. For other isolates, only DNA was available.
Accession numbers for the sequences analyzed in this study.
Restriction fragment length polymorphism of the rrf-rrl amplicons.
The small ribosomal spacer rrf-rrl was amplified using primers 1 and 2, as previously described (28). The analysis of the size of fragments obtained after restriction by MseI and in some cases by DraI allowed us first to identify Borrelia species and then to point out any atypical pattern.
PFGE.
Previously described procedures were used for the preparation of high-molecular-weight genomic DNAs and for PFGE (4, 9). Rare-cutting endonucleases SacII, BssHII, and MluI were used to differentiate isolates. Lambda concatemers (monomer size, 48.5 kbp) purchased from New England Biolabs, Beverly, Mass., were used as a size control. All fragments on the gels resulting from restrictions were transformed as binary data, where each fragment was scored as 1 (present) or 0 (absent). These data yielded a binary matrix that was used to draw a phylogenetic tree.
Analysis of ospC diversity by SSCP-PCR.
To screen for the genetic diversity of the ospC gene, we used a single-strand conformation polymorphism (SSCP) analysis. SSCP is a screening method based on the secondary structure of a single-stranded DNA fragment. A 277-bp fragment in the ospC gene was amplified by PCR using the primer set F-SC3 and R-OspC9 under the conditions previously described (19). Briefly, after denaturation, PCR samples were loaded on a polyacrylamide gel. Electrophoresis was conducted in a temperature-controlled electrophoresis system (GenePhor; Amersham Pharmacia Biotech) at 6°C with a first run at 600 V, 25 mA, and 15 W for 10 min and then at 600 V, 37 mA, and 21 W for 2.5 h. Gels were revealed by silver staining according to the manufacturer's instructions (Plus One DNA silver staining kit; Amersham Pharmacia Biotech).
Sequencing and phylogenetic analysis of sequences.
Intergenic spacer PCR products were sequenced by Genome Express (Meylan, France). Amplification products from the ospA gene were sequenced using the BigDye Terminator cycle sequencing kit (Applied Biosystems) as described in reference 12. Primers used in this study were direct and reverse primers 1 and 2, as previously described (12).
Sequences were aligned by using Clustal V software and manually by using VSM software and then analyzed by either the unweighted pair group with mathematical average (UPGMA) (36) or the neighbor-joining (NJ) method (34). Phylogenetic trees were drawn with Mega software (18). Sequences of North African B. lusitaniae isolates were compared to additional sequences from B. lusitaniae and from other species available in data banks.
RESULTS
The isolates obtained from I. ricinus ticks collected in North Africa and analyzed in this study are shown in Table 1. Additional DNAs were used for determination of the sole rrf-rrl spacer sequence; they appear in Fig. 1.
FIG. 1.
UPGMA-rooted tree obtained with MEGA software from the ribosomal rrf-rrl spacer sequences. Sequences issued from data banks are followed by the sequence accession number. AF, Africa; E, Europe; AS, Asia; Mo, Morocco; Tu, Tunisia; Po, Portugal; Tk, Turkey; Sp, Spain; Mol, Moldavia.
rrf-rrl ribosomal spacer.
The analysis of MseI restriction patterns from amplification products of the rrf-rrl spacer was performed to identify, at a species level, 514 Borrelia DNA isolated from I. ricinus ticks collected in North Africa in 2000 to 2003. Only 10 DNAs belonged to either B. burgdorferi sensu stricto (n = 3), B. garinii (n = 6), or B. valaisiana (n = 1). The remaining 504 DNAs belonged to the species B. lusitaniae. PCR products that exhibited atypical patterns were sequenced, as were some randomly chosen products whose patterns were similar to that of PotiB2T. Thus, sequences of 79 PCR products were determined, 44 from strains isolated in Tunisia and 35 that originated from strains isolated in Morocco.
Six different restriction patterns were observed, as shown in Table 2. A large majority of MseI patterns (n = 456) were similar to that of the type strain PotiB2T. Sequences of strains exhibiting a similar restriction pattern differed from the PotiB2T sequence by only one or two nucleotide deletions. Interestingly, the MseI and DraI patterns of strain M3-22 were indistinguishable from those of strain PotiB2T on the gels. However, sequencing revealed an insertion of 15 nucleotides containing an MseI and DraI restriction site leading to an additional 16-bp-long fragment not visible on the gels. The remaining sequence was identical to that of strain PotiB2T. Consequently, this pattern may be underscored when no sequencing is performed. Four Tunisian strains were identical to strain TD12 (44). Two previously undescribed patterns were identified and named MT26 and MA3-13 (Table 2). Five Moroccan strains exhibited the MT26 pattern, and two strains exhibited the MA3-13 pattern. These patterns have been confirmed by sequencing as belonging to B. lusitaniae, close to PotiB2T. Only 11 strains showed the typical pattern of strain PotiB3, one of the first genotypes of B. lusitaniae described in Portugal.
TABLE 2.
Restriction fragment analysis of rrl-rrf amplicons
| Genotype | No. of DNA belonging to each genotype | Amplicon size (bp) | MseI restriction pattern (bp) | DraI restriction pattern (bp) |
|---|---|---|---|---|
| PotiB2 | 456 | 257 | 107, 82, 39, 29 | 145, 83, 29 |
| PotiB3 | 11 | 255 | 107, 80, 52, 16 | 158, 81, 16 |
| MT26 | 5 | 241 | 107, 95, 39 | 145, 96 |
| MA3-13 | 2 | 246 | 107, 80, 30, 29 | 136, 81, 29 |
| M3-22 | 1 | 271 | 107, 81, 38, 29, 16 | 144, 82, 29, 16 |
| TD12 | 4 | 255 | 107, 80, 39, 17, 12 | 145, 81, 29 |
A phylogenetic tree was constructed from sequences of B. lusitaniae isolates that originated from Morocco and Tunisia (Fig. 1). Whatever the method used (UPGMA or NJ), the sequences scattered in two major clusters, although they were separated by only short genetic distances. One cluster contained sequences related to the PotiB3 genotype. Despite a restriction pattern close to that of the PotiB2T strain, the sequence of M3-22 appeared phylogenetically closer to the PotiB3 sequence than to that of PotiB2T. The second cluster included sequences related to PotiB2T. In this cluster, the higher diversity was observed among sequences from Portuguese strains obtained from data banks. In contrast, a very low level of diversity was observed among sequences from North African strains. Sixty-three sequences were included in the major branch (Fig. 1), and these segregated with two sequences from Portuguese strains, one sequence from a Spanish strain, and one from a Moldavian strain. All these sequences differed from that of PotiB2T and from each other by only one or two nucleotide substitutions. Four strains (pattern MT26) shared an identical sequence to that of PotiB2T except a deletion of 16 bp, including an MseI restriction site. Therefore, these sequences segregated in a separate branch together with PotiB2T. The MA3-13 sequence totally matched with one sequence of a Portuguese strain available in data banks. The two sequences were located on a separated branch in the PotiB2T cluster.
Whole genome analyzed by PFGE.
Thirty-three North African B. lusitaniae isolates were analyzed by PFGE after DNA restriction by three enzymes, SacII, BssHII (Fig. 2), and MluI, and compared to the Portuguese strains PotiB2T and PotiB3. Only discrete restriction fragments of >50 kb were included in the analysis.
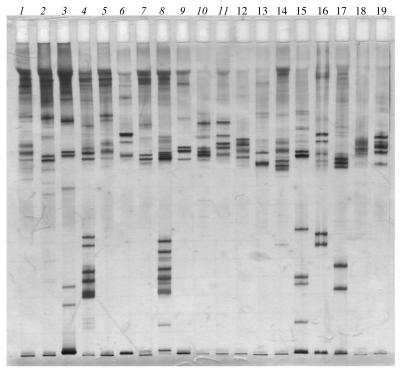
FIG. 2.
BssHII macrorestriction patterns of B. lusitaniae strains obtained with the Bio-Rad apparatus with the pulse time ramped from 2 to 70 s for 30 h. (A) Moroccan strains. The lanes contained lambda concatemers (lanes 1 and 11), PotiB2 (lane 2), MT1 (lane 3), MT7 (lane 4), MT18 (lane 5), MT20 (lane 6), MT24 (lane 7), MT25 (lane 8), MT27 (lane 9), and PotiB3 (lane 10). (B) Tunisian strains. The lanes contained lambda concatemers (lanes 1 and 13), PotiB2 (lane 2), PotiB3 (lane 3), TT840 (lane 4), TT867 (lane 5), TT894 (lane 6), TT899 (lane 7), TT908 (lane 8), TT916 (lane 9), TT918 (lane 10), TT919 (lane 11), and TT920 (lane 12).
All B. lusitaniae strains studied in this work were resolved into three specific patterns after restriction by MluI, whereas restriction by either SacII or BssHII afforded a greater polymorphism. However, a majority of strains were distributed into a few macrorestriction patterns. Even though nine and seven different patterns were observed after restriction by SacII and BssHII, respectively, 24 (72.7%) and 27 (81.8%) North African strains scattered on two patterns after restriction by each enzyme. Most strains presented a similar pattern as the type strain PotiB2T.
Although restriction by SacII led to the higher polymorphism, it did not allow recognition of the PotiB3 genotype, which can be easily distinguished from PotiB2 after restriction by BssHII and MluI.
All PFGE data yielded a matrix of 35 strains by 25 characters that was subjected to a phylogenetic analysis by NJ and UPGMA methods. The pulsotype PotiB3 clustered separately from other B. lusitaniae pulsotypes resolved in two large divisions. The largest division contained isolates that segregated with PotiB2T (Fig. 3). As shown in Fig. 3, 33 Moroccan and Tunisian strains were equally distributed into 19 alleles.
FIG. 3.
Phylogenetic analysis of polymorphism after DNA restriction with MluI, BssHII, and SacII and resolution by PFGE. All fragments were transformed into a binary matrix used to generate a tree by the UPGMA method.
ospA sequences.
Sixteen partial ospA sequences have been determined, 10 from Moroccan strains and 6 from Tunisian strains. Sequences from B. lusitaniae clustered separately from other B. burgdorferi sensu lato species (data not shown). Within the B. lusitaniae cluster, there was no significant clustering of isolates from Tunisia or Morocco, whereas the three Portuguese strains PotiB1, PotiB2T, and PotiB3 were located on an individualized branch.
ospC gene mobility classes.
Thirty-three DNAs, 16 from ticks collected in Morocco and 17 from ticks collected in Tunisia, were analyzed by SSCP-PCR. A consistent diversity was found. With the exception of two couples of Tunisian strains that each shared a similar pattern (data not shown), each DNA studied was characterized by a specific SSCP pattern (Fig. 4).
FIG. 4.
ospC SSCP patterns of Moroccan B. lusitaniae strains. Lane 1, MT2; lane 2, MT4; lane 3, MT8; lane 4, MT9; lane 5, MT10; lane 6, MT11; lane 7, MT12; lane 8, MT14; lane 9, MT16; lane 10, B. lusitaniae PotiB1; lane 11, B. lusitaniae PotiB2T; lane 12, B. lusitaniae PotiB3; lane 13, MT17; lane 14, MT20; lane 15, MT22; lane 16, MT24; lane 17, MT27; lane 18, MT13, lane 19, MT26.
DISCUSSION
In contrast to the other B. burgdorferi sensu lato species distributed throughout Europe, B. lusitaniae only occurs sporadically in different states, mainly in Central Europe. Since the vector I. ricinus is largely spread over Europe, the reason for the restricted geographic distribution of B. lusitaniae remains unclear. However, in a study in Portugal, B. lusitaniae was the sole B. burgdorferi sensu lato species observed in I. ricinus ticks (6). Similarly, it is by far the most predominant species isolated from ticks in North Africa (35, 44, 46). A large diversity of the small rrf-rrl spacer was reported among the Portuguese strains (6). In this context, we were interested in analyzing the genetic diversity of B. lusitaniae in North Africa.
Most authors have analyzed the global genetic variability of B. burgdorferi sensu lato by the analysis of the macrorestriction polymorphism with one enzyme (MluI). If we considered the MluI restriction polymorphism, only three patterns were recorded among 35 strains. By using three enzymes, we observed a global diversity similar to that reported for B. burgdorferi sensu stricto or B. garinii after restriction by only one enzyme, confirming a low diversity (4, 9, 10, 23). Our results did not evidence any clear clustering among strains isolated in Tunisia or Morocco (Fig. 3).
In order to obtain a higher level of discrimination among B. lusitaniae strains, we also focused on two loci, ospA and ospC. The former is subjected to an adaptive pressure, whereas the latter is subjected to a diversifying frequency-dependent selection (2, 8), with these two different mechanisms leading to distinct ranges of variability. The ospC diversity should be due to an immune pressure from locally prominent hosts, preventing a reinfection by an antigenically related strain. Therefore, whatever the host or vector spectrum, the diversity is expected to be high. This was confirmed for B. lusitaniae in North Africa. The genetic variability of the ospC gene of pathogenic B. burgdorferi sensu lato strains has been extensively studied (14, 21, 37, 39). The diversity of the ospC gene is mostly acquired by lateral transfer between species or within species. However, given both the restricted geographic distribution of B. lusitaniae and the scarcity of isolates from other species in North Africa, incorporation of sequences from other B. burgdorferi sensu lato species is unlikely. To clarify this point, it would be worthwhile to extend the study to B. lusitaniae strains from other geographic areas. ospA gene sequence analyses allow identification of species within B. burgdorferi sensu lato (23, 42, 43), except for B. valaisiana. It is noteworthy that the ospA sequences from Portuguese Poti strains clustered separately, although close to North African sequences (data not shown). No precise correlation could be established between segregation of ospA and macrorestriction patterns. The relative level of conservation of ospA sequences from B. lusitaniae could be attributed to the restricted geographic extension and to the interaction with only one vector species and possibly one predominant reservoir host. This fact contrasts with the diversity reported for B. valaisiana, which is more widely distributed and is associated with several tick species, requiring adaptation to a broad range of vectors (12, 22, 24, 32, 40).
Our analysis of the restriction polymorphism of the small rrf-rrl spacer emphasized a high monomorphism that was confirmed by the conservation of sequences within B. lusitaniae isolates from North Africa. Fourteen alleles were recorded for the whole B. lusitaniae sequences. Among these 14 alleles, 11 contained 14 available sequences from Portuguese isolates, whereas only 7 alleles contained 79 sequences from North African isolates. Only three alleles present among North African isolates were specific to this continent and had no counterpart in Portugal. Therefore, a considerable rrf-rrl sequence heterogeneity seems to characterize Portuguese strains, in contrast to North African strains. However, De Michelis et al. (6) reported that different alleles were not equally represented, with 50% of Portuguese strains belonging to allele I (strain GT058). It is noteworthy that 80.7% of North African sequences also belonged to this allele (Fig. 1), as did the Moldavian strain Ir345 (20, 29). The description of three new alleles from North African strains does not significantly increase the global genetic diversity previously known for the species B. lusitaniae on this locus. Given the large number of sequences determined, it is likely that the global diversity of the species has been assessed. It is noteworthy that no sequence from strains isolated in North Africa fell in the same allele as strain PoHL isolated from a Portuguese patient.
It must be emphasized that the genetic heterogeneity involving the ribosomal spacer was essentially acquired by insertion or deletion of short DNA fragments (data not shown). Very few mutations were observed along the sequences. If we consider that this locus is not subjected to any selection pressure and mostly reflects the molecular evolutionary clock, we could hypothesize that either North African B. lusitaniae strains recently evolved from a common ancestor or only a few prevalent genotypes have successfully settled in North Africa. Considering that the whole diversity of the species is present in Portugal, it is likely that B. lusitaniae has evolved for a considerable time in Portugal and has been recently introduced in North Africa. Therefore, the analysis of a chromosomal locus makes obvious a founding event leading to the recent emergence of some North African clones sampled from the Portuguese diversity.
Our findings with a large population of B. lusitaniae strains from North Africa support the conclusion that a high proportion of this population belongs to a limited number of closely related genotypes. This is in accordance with the clonal structure of the Borrelia population as previously demonstrated (7). Some cases of Lyme borreliosis have been reported in Tunisia, based on clinical and serological features (1), and two cases of facial palsy have been reported in Morocco (27). Moreover, a recent study indicated that the inoculation of B. lusitaniae into susceptible mice induced pathological features (45). Finally, B. lusitaniae was recently isolated, for the first time, from a chronic skin lesion in a Portuguese patient (5). If this species is actually pathogenic for humans, the risk of Lyme borreliosis must be underlined in North Africa. However, the pathogenicity could be restricted to distinct clones which have not yet settled in North Africa. Additional studies are needed to clarify the evolutionary events leading to the emergence of a restricted population of bacteria in a given geographical area.
Acknowledgments
This work was supported by the European Commission, grant ICA3-CT-2000-30009, the Swiss Federal Office of Education and Science (99.0695), and the Roche Research Foundation (Switzerland). This work is part of the Ph.D. thesis of Hend Younsi.
We thank K. Dellagi and M. Hassar, Directors of the Pasteur Institute of Tunisia and Morocco, respectively, for their constant support. The language of the manuscript was revised by Iain Old.
REFERENCES
- 1.Aoun, K., A. Kechrid, N. Lagha, A. Zarrouk, and N. Bouzouaia. 1998. La maladie de Lyme en Tunisie, résultats d'une étude clinico-sérologique (1992-1996). Cah. Santé 8:98-100. [PubMed] [Google Scholar]
- 2.Baranton, G., G. Seinost, G. Theodore, D. Postic, and D. Dykhuizen. 2001. Distinct levels of genetic diversity of Borrelia burgdorferi are associated with different aspects of pathogenicity. Res. Microbiol. 152:149-156. [DOI] [PubMed] [Google Scholar]
- 3.Barral, M., A. L. Garcia-Perez, R. A. Juste, A. Hurtado, R. Escudero, R. E. Sellek, and P. Anda. 2002. Distribution of Borrelia burgdorferi sensu lato in Ixodes ricinus (Acari:Ixodidae) ticks from the Basque Country, Spain. J. Med. Entomol. 39:177-184. [DOI] [PubMed] [Google Scholar]
- 4.Belfaiza, J., D. Postic, E. Bellenger, G. Baranton, and I. Saint Girons. 1993. Genomic fingerprinting of Borrelia burgdorferi sensu lato by pulsed-field gel electrophoresis. J. Clin. Microbiol. 31:2873-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collares-Pereira, M., S. Couceiro, I. Franca, K. Kurtenbach, S. M. Schäfer, L. Vitorino, L. Gonçalves, S. Baptista, M. L. Viera, and C. Cunha. 2004. First isolation of Borrelia lusitaniae from a human patient. J. Clin. Microbiol. 42:1316-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Michelis, S., H. S. Sewell, M. Collares-Pereira, M. Santos-Reis, L. M. Schouls, V. Benes, E. C. Holmes, and K. Kurtenbach. 2000. Genetic diversity of Borrelia burgdorferi sensu lato in ticks from mainland Portugal. J. Clin. Microbiol. 38:2128-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dykhuizen, D. E., and G. Baranton. 2001. Borrelia burgdorferi, a (somewhat) clonal bacterial species. Trends Microbiol. 9:472. [DOI] [PubMed] [Google Scholar]
- 8.Dykhuizen, D. E., and G. Baranton. 2001. The implications of a low rate of horizontal transfer in Borrelia. Trends Microbiol. 9:344-350. [DOI] [PubMed] [Google Scholar]
- 9.Foretz, M., D. Postic, and G. Baranton. 1997. Phylogenetic analysis of Borrelia burgdorferi sensu stricto by arbitrarily primed PCR and pulsed-field gel electrophoresis. Int. J. Syst. Bacteriol. 47:11-18. [DOI] [PubMed] [Google Scholar]
- 10.Fukunaga, M., and Y. Takahashi. 1994. Pulsed field gel electrophoresis analysis of Borrelia burgdorferi sensu lato isolated in Japan and taxonomic implications with Lyme disease spirochetes. Microbiol. Immunol. 38:747-751. [DOI] [PubMed] [Google Scholar]
- 11.Gern, L., C. M. Hu, E. Kocianova, V. Vyrostekova, and J. Rehacek. 1999. Genetic diversity of Borrelia burgdorferi sensu lato isolates obtained from Ixodes ricinus ticks collected in Slovakia. Eur. J. Epidemiol. 15:665-669. [DOI] [PubMed] [Google Scholar]
- 12.Godfroid, E., C. M. Hu, P.-F. Humair, A. Bollen, and L. Gern. 2003. PCR-reverse line blot typing method underscores the genomic heterogeneity of Borrelia valaisiana species and suggests its potential involvement in Lyme disease. J. Clin. Microbiol. 41:3690-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Güner, E. C., N. Hashimoto, N. Takada, K. Kaneda, Y. Imai, and T. Masuzawa. 2003. First isolation and characterization of Borrelia burgdorferi sensu lato strains from Ixodes ricinus ticks in Turkey. J. Med. Microbiol. 52:807-813. [DOI] [PubMed] [Google Scholar]
- 14.Jauris-Heipke, S., G. Liegl, V. Preac-Mursic, D. Röbler, E. Schwab, E. Soutschek, G. Will, and B. Wilske. 1995. Molecular analysis of genes encoding outer surface protein C (OspC) of Borrelia burgdorferi sensu lato: relationship to ospA genotype and evidence of lateral gene exchange of ospC. J. Clin. Microbiol. 33:1860-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jouda, F., M. Crippa, J. L. Perret, and L. Gern. 2003. Distribution and prevalence of Borrelia burgdorferi sensu lato in Ixodes ricinus ticks of Canton Ticino (Switzerland). Eur. J. Epidemiol. 18:907-912. [DOI] [PubMed] [Google Scholar]
- 16.Jouda, F., J. L. Perret, and L. Gern. 2004. Density of questing Ixodes ricinus nymphs and adults infected by Borrelia burgdorferi sensu lato in Switzerland: spatio-temporal pattern at a regional scale. Vector Borne Zoon. Dis. 4:23-32. [DOI] [PubMed] [Google Scholar]
- 17.Jouda, F., J. L. Perret, and L. Gern. 2004. Ixodes ricinus density, and distribution and prevalence of Borrelia burgdorferi sensu lato infection along an altitudinal gradient. J. Med. Entomol. 41:162-170. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, S., K. Tamura, and N. Masatoshi. 1993. MEGA: molecular evolutionary genetics analysis, version 1.01. The Pennsylvania State University, University Park, Pa.
- 19.Lagal, V., D. Postic, E. Ruzic-Sabljic, and G. Baranton. 2003. Genetic diversity among Borrelia strains determined by single-strand conformation polymorphism analysis of the ospC gene and its association with invasiveness. J. Clin. Microbiol. 41:5059-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Flèche, A., D. Postic, K. Girardet, O. Péter, and G. Baranton. 1997. Characterization of Borrelia lusitaniae sp. nov. by 16S ribosomal DNA sequence analysis. Int. J. Syst. Bacteriol. 47:921-925. [DOI] [PubMed] [Google Scholar]
- 21.Livey, I., C. P. Gibbs, R. Schuster, and F. Dorner. 1995. Evidence for lateral transfer and recombination in OspC variation in Lyme disease Borrelia. Mol. Microbiol. 18:257-269. [DOI] [PubMed] [Google Scholar]
- 22.Masuzawa, T., T. Fukui, M. Miyake, H. B. Oh, M. K. Cho, W. H. Chang, Y. Imai, and Y. Yanagihara. 1999. Determination of members of a Borrelia afzelii-related group isolated from Ixodes nipponensis in Korea as Borrelia valaisiana. Int. J. Syst. Bacteriol. 49:1409-1415. [DOI] [PubMed] [Google Scholar]
- 23.Mathiesen, D. A., J. H. Oliver, Jr., C. P. Kolbert, E. D. Tullson, B. J. B. Johnson, G. L. Campbell, P. D. Mitchell, K. D. Reed, S. R. Telford III, J. F. Anderson, R. S. Lane, and D. H. Persing. 1997. Genetic heterogeneity of Borrelia burgdorferi in the United States. J. Infect. Dis. 175:98-107. [DOI] [PubMed] [Google Scholar]
- 24.Miyamoto, K., and T. Masuzawa. 2002. Ecology of Borrelia burgdorferi sensu lato in Japan and East Asia, p. 201-222. In O. Kahl, J. Gray, R. S. Lane, and G. Stanek (ed.), Lyme borreliosis: biology, epidemiology and control. CABI Publishing, Wallingford, United Kingdom.
- 25.Mizak, B., and J. Krol. 2000. Analysis of Polish isolates of Borrelia burgdorferi by amplification of rrf (5S)-rrl (23S) intergenic spacer. Bull. Vet. Inst. Pulawy 44:147-154. [Google Scholar]
- 26.Nuncio, M. S., O. Péter, M. J. Alves, F. Bacellar, and A. R. Filipe. 1993. Isolamento e caracterizaçao de Borrélias de Ixodes ricinus L. em Portugal. Rev. Portuguesa Doenças Infecc. 16:175-179. [Google Scholar]
- 27.Ouhabi, H., I. Slassi, M. El Aloui-Frais, R. Mossadaq, M. Yahyaoui, and T. Chkili. 1994. Paralysie faciale et maladie de Lyme. Arch. Inst. Past. Maroc. 9:51-55. [Google Scholar]
- 28.Postic, D., M. V. Assous, P. A. D. Grimont, and G. Baranton. 1994. Diversity of Borrelia burgdorferi sensu lato evidenced by restriction fragment length polymorphism of rrf (5S) rrl (23S) intergenic spacer amplicons. Int. J. Syst. Bacteriol. 44:743-752. [DOI] [PubMed] [Google Scholar]
- 29.Postic, D., E. Korenberg, N. Gorelova, Y. V. Kovalevski, E. Bellenger, and G. Baranton. 1997. Borrelia burgdorferi sensu lato in Russia and neighboring countries: high incidence of mixed isolates. Res. Microbiol. 148:691-702. [DOI] [PubMed] [Google Scholar]
- 30.Richter, D., D. B. Schlee, and F. R. Matuschka. 2003. Relapsing fever-like spirochetes infecting European vector tick of Lyme disease agent. Emerg. Infect. Dis. 9:697-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rijpkema, S. G. T., D. J. Tazelaar, M. J. C. H. Molkenboer, G. T. Noordhoek, G. Plantinga, L. M. Schouls, and J. F. P. Schellekens. 1997. Detection of Borrelia afzelii, Borrelia burgdorferi sensu stricto, Borrelia garinii and group VS116 by PCR in skin biopsies of patients with erythema migrans and acrodermatitis chronica atrophicans. Clin. Microbiol. Infect. 3:109-116. [DOI] [PubMed] [Google Scholar]
- 32.Ryffel, K., O. Péter, E. Dayer, A. Bretz, and E. Godfroid. 2003. OspA heterogeneity of Borrelia valaisiana confirmed by phenotypic and genotypic analyses. BMC Infect. Dis. 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryffel, K., O. Péter, B. Rutti, A. Suard, and E. Dayer. 1999. Scored antibody reactivity determined by immunoblotting shows an association between clinical manifestations and presence of Borrelia burgdorferi sensu stricto, B. garinii, B. afzelii, and B. valaisiana in humans. J. Clin. Microbiol. 37:4086-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 35.Sarih, M., F. Jouda, L. Gern, and D. Postic. 2003. First isolation of Borrelia burgdorferi sensu lato from ticks in Morocco. Vector Borne Zoon. Dis. 3:133-139. [DOI] [PubMed] [Google Scholar]
- 36.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. Freeman, San Francisco, Calif.
- 37.Theisen, M., B. Frederiksen, A. M. Lebech, J. Vuust, and K. Hansen. 1993. Polymorphism in ospC gene of Borrelia burgdorferi and immunoreactivity of OspC protein: implications for taxonomy and for use of OspC protein as a diagnostic antigen. J. Clin. Microbiol. 31:2570-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, G., A. P. van Dam, A. Le Flèche, D. Postic, O. Péter, G. Baranton, R. de Boer, L. Spanjaard, and J. Dankert. 1997. Genetic and phenotypic analysis of Borrelia valaisiana sp. nov. (Borrelia genomic groups VS116 and M19). Int. J. Syst. Bacteriol. 47:926-932. [DOI] [PubMed] [Google Scholar]
- 39.Wang, G. Q., A. P. van Dam, and J. Dankert. 1999. Evidence for frequent ospC gene transfer between Borrelia valaisiana sp. nov. and other Lyme disease spirochetes. FEMS Microbiol. Lett. 177:289-296. [DOI] [PubMed] [Google Scholar]
- 40.Wang, G. Q., A. P. van Dam, and J. Dankert. 2000. Two distinct ospA genes among Borrelia valaisiana strains. Res. Microbiol. 151:325-331. [DOI] [PubMed] [Google Scholar]
- 41.Wang, G. Q., A. P. van Dam, I. Schwartz, and J. Dankert. 1999. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin. Microbiol. Rev. 12:633-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Will, G., S. Jauris-Heipke, E. Schwab, U. Busch, D. Rossler, E. Soutschek, B. Wilske, and V. Preac-Mursic. 1995. Sequence analysis of ospA genes shows homogeneity within Borrelia burgdorferi sensu stricto and Borrelia afzelii strains but reveals major subgroups within the Borrelia garinii species. Med. Microbiol. Immunol. 184:73-80. [DOI] [PubMed] [Google Scholar]
- 43.Wilske, B., V. Preac-Mursic, U. B. Göbel, B. Graf, S. Jauris, E. Soutschek, E. Schwab, and G. Zumstein. 1993. An OspA serotyping system for Borrelia burgdorferi based on reactivity with monoclonal antibodies and OspA sequence analysis. J. Clin. Microbiol. 31:340-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Younsi, H., D. Postic, G. Baranton, and A. Bouattour. 2001. High prevalence of Borrelia lusitaniae in Ixodes ricinus ticks in Tunisia. Eur. J. Epidemiol. 17:53-56. [DOI] [PubMed] [Google Scholar]
- 45.Zeidner, N. S., M. S. Nuncio, B. S. Schneider, L. Gern, J. Piesman, O. Brandao, and A. R. Filipe. 2001. A Portuguese isolate of Borrelia lusitaniae induces disease in C3H/HeN mice. J. Med. Microbiol. 50:1055-1060. [DOI] [PubMed] [Google Scholar]
- 46.Zhioua, E., A. Bouattour, C. M. Hu, M. Gharbi, A. Aeschliman, H. S. Ginsberg, and L. Gern. 1999. Infection of Ixodes ricinus (Acari:Ixodidae) by Borrelia burgdorferi sensu lato in North Africa. J. Med. Entomol. 36:216-218. [DOI] [PubMed] [Google Scholar]