Figure 3.
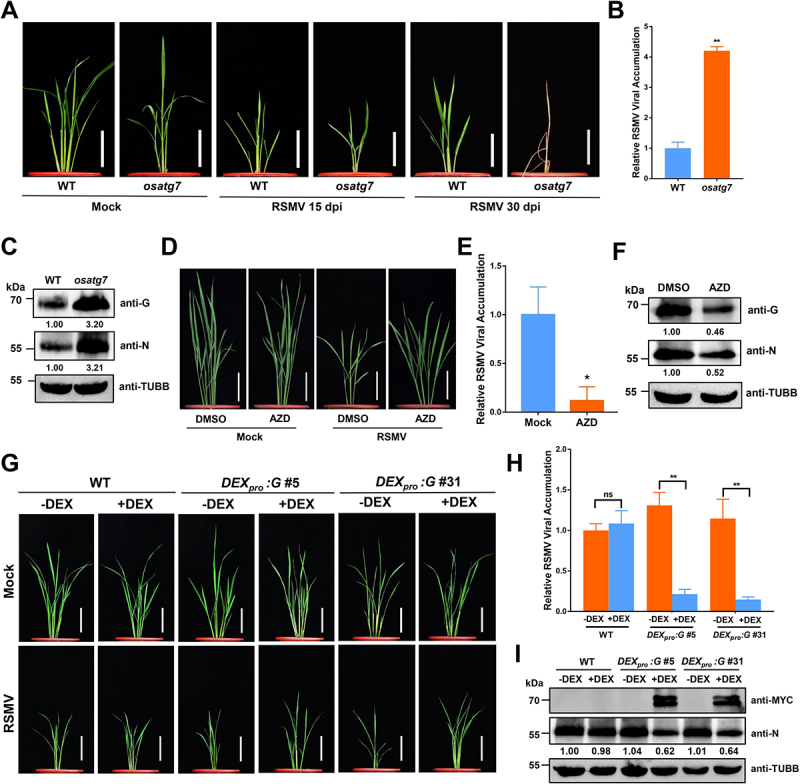
Autophagy plays an antiviral role during RSMV infection. (A) the symptoms of RSMV infection in osatg7 mutant and WT plants. The four-leaf stage plants were inoculated by viruliferous leafhopper and the photographs were taken at 15 and 30 dpi. Bars: 10 cm. (B) qRT-PCR analysis of RSMV viral RNA accumulation in osatg7 mutant and WT plants. Total RNAs were extracted from the RSMV infected leaves at 15 dpi. Values represent the mean relative to the WT plants (n = 3 biological replicates) and were normalized with OsEf1α as an internal reference. Student’s t test was used for analyses (*P<.01). (C) Western blot analysis of RSMV viral proteins accumulation in osatg7 mutant and WT plants. Total proteins were extracted from the RSMV infected leaves at 15 dpi, and were detected by specific antibodies against RSMV glycoprotein (anti-G) and nucleocapsid protein (anti-N). The relative protein band intensity was normalized against that of TUBB/tubulin. (D) the symptoms of RSMV infection in rice plants treated with DMSO or AZD. The plants were inoculated by viruliferous leafhopper and 10 μM AZD or DMSO as solvent control were treated for 3 h each by root soaking at 1, 4, 7, and 10 dpi. The photographs were taken at 30 dpi. Bars: 10 cm. (E) qRT-PCR analysis of RSMV viral RNA accumulation in rice plants treated with DMSO or AZD. Total RNAs were extracted from the RSMV infected leaves at 15 dpi. Values represent the mean relative to the WT plants (n = 3 biological replicates) and were normalized with OsEf1α as an internal reference. Student’s t test was used for analyses (**P<.05). (F) Western blot analysis of RSMV viral proteins accumulation in rice plants treated with DMSO or AZD. Total proteins were extracted from the RSMV infected leaves at 15 dpi, and were detected by specific antibodies against RSMV glycoprotein (anti-G) and nucleocapsid protein (anti-N). The relative protein band intensity was normalized against that of TUBB/tubulin. (G) the symptoms of RSMV infection in WT and DEXpro:G transgenic rice plants treated with or without DEX. The plants were inoculated by viruliferous leafhopper and then 50 μM DEX or solvent control was sprayed on the leaf surface at 1 dpi. The photographs were taken at 30 dpi. Bars: 10 cm. (H) qRT-PCR analysis of RSMV viral RNA accumulation in WT and DEXpro:G transgenic rice plants treated with or without DEX. Total RNAs were extracted from the RSMV infected leaves at 15 dpi. Values represent the mean relative to the WT plants (n = 3 biological replicates) and were normalized with OsEf1α as an internal reference. Two-way ANOVA followed by multiple-comparisons Tukey’s test was used for analyses (**P < 0.01). ns, not significant. (I) Western blot analysis of RSMV viral proteins accumulation in WT and DEXpro:G transgenic rice plants treated with or without DEX. Total proteins were extracted from the RSMV infected leaves at 15 dpi, and were detected by specific antibodies against MYC-tag (transgenic expressed glycoprotein fused with MYC tag) and RSMV nucleocapsid protein (anti-N). The relative protein band intensity was normalized against that of TUBB/tubulin.
