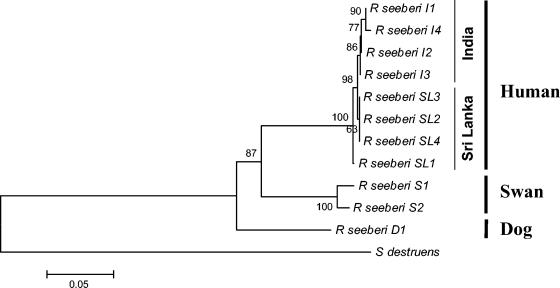
FIG. 1.
Phylogenetic relationship between the ITS1, 5.8S, and ITS2 sequences of eight human strains of R. seeberi from India (four strains; I1 to I4) and Sri Lanka (four strains; SL1 to SL4), one strain of R. seeberi from a dog (D1), and two strains from swans (S1 and S2). Multiple hit correction and a 1,000-bootstrap-resampled data set were used to assess branch support. The ITS sequences of R. seeberi isolated from humans are all clustered in a solid group. In this group, the strains from India and Sri Lanka grouped in two poorly supported subgroups. In contrast, the two R. seeberi strains from swans formed a strongly supported sister group to the human strains (100% bootstrap) and with the inclusion of R. seeberi from a dog, a well-supported sister group to the dog (87% bootstrap). Numbers above the branches are percentages of bootstrap-resampled data sets obtained by neighbor-joining. The ITS sequences of S. destruens were used as out-group. The scale bar represents evolutionary distance in substitutions per nucleotide.