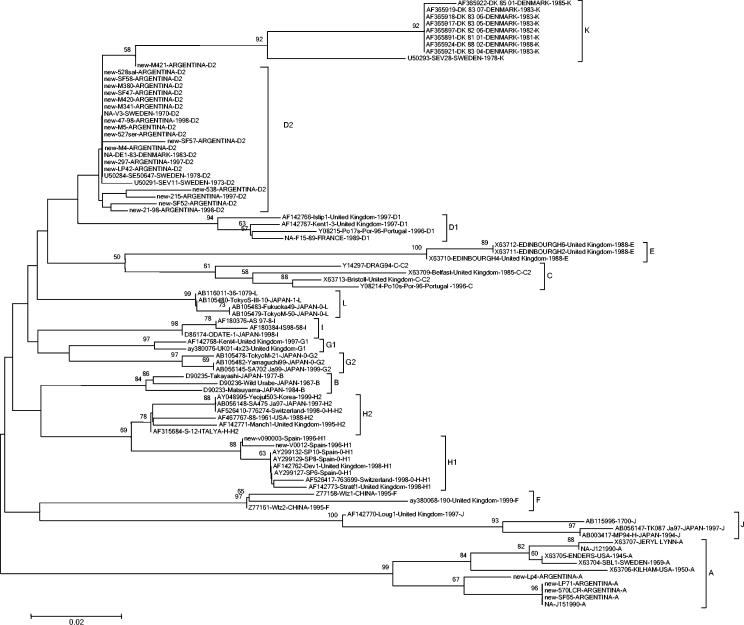
FIG. 1.
The set of nucleotide sequences was aligned with Clustal W. Phylogenetic analysis was performed using the Kimura two-parameter model as a model of nucleotide substitution and using the neighbor-joining method to reconstruct the phylogenetic tree (MEGA version 2.1 software package). The statistical significance of the phylogenies constructed was estimated by bootstrap analysis with 1,000 pseudoreplicate data sets. For clarity, only a subset of the database is shown.