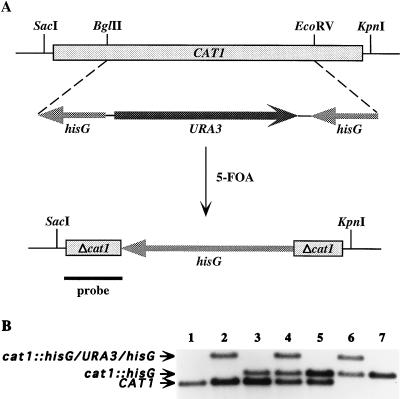
FIG. 2.
Sequential disruption of the CAT1 gene of C. albicans by using the Ura-blaster technique. (A) Partial restriction map of the CAT1 allele before and after regeneration of the ura3 genetic marker by 5-FOA selection. The plasmid used for the gene disruption construct encompassed the entire 2,036 bp containing the entire 1,461-kb CAT1 ORF, indicated by the boxed and shaded rectangle at the top. Restriction sites BglII, EcoRV, KpnI, and SacI are indicated. A 1,351-bp BglII-EcoRV fragment of CAT1 was replaced by the his-URA-his construct; this is shown below the CAT1 ORF. The his-URA-his construct consisted of URA3 flanked by a hisG insert at each end. Beneath this construct are shown the final transformants selected by 5-FOA, which contained the disrupted CAT1 gene, missing the 1,351-bp BglII-EcoRV fragment and retaining one copy of hisG (1,149 bp) flanked by small fragments of the disrupted CAT1 gene (Δcat1). The solid black line at the bottom represents the location of a portion of CAT1 used as a 32P-labeled probe for Southern analysis. It is important to note that the lengths of each of the fragments are not drawn to scale. (B) Southern analysis of the parental, heterozygous, and homozygous mutants after digestion with KpnI/SacI, followed by hybridization using a fragment of CAT1 as a probe. Lanes: 1, CAI4 parental DNA; 2 to 7, pre- and post-5-FOA isolates after the first (lanes 2 and 3), second (lanes 4 and 5), and third (lanes 6 and 7) rounds of transformation. Not shown in this figure, Southern analysis after hybridization with a hisG probe confirmed that hisG was present in each of the cat1::hisG bands (lanes 3 to 7) and cat1::hisG/URA3/hisG bands (lanes 2, 4, and 6) but not with any of the CAT1 bands (lanes 1 to 5).