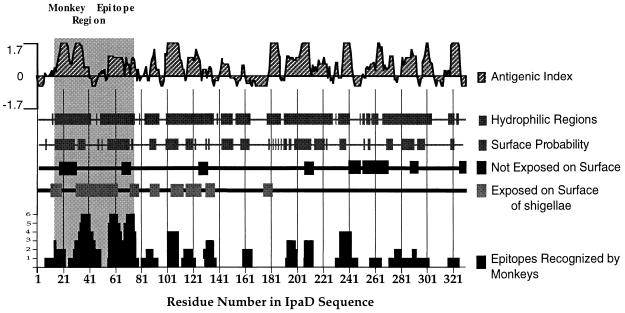
FIG. 3.
Relationship of the predicted secondary structure of IpaD with experimentally identified epitopes. By using the deduced amino acid sequence of IpaD (28) and the PROTEAN analysis software, the antigenic index, hydrophilic regions, and surface probability were calculated. Peptide epitopes exposed on the surface or not accessible to antibodies are indicated. Surface epitopes were identified with rabbit sera or MAbs 2A9 and 16F8 made against IpaD peptides or rIpaD. Peptide epitopes recognized by 10 different monkeys challenged with S. flexneri 2a are also plotted. The height of the column corresponds to the number of monkeys (vertical axis, 1 to 6) recognizing a particular residue within a peptide epitope. All secondary structure and epitope data are plotted against the amino acid residue number in the IpaD sequence; the highly immunogenic region between amino acid residues 14 and 77 is also indicated (grey shaded area).