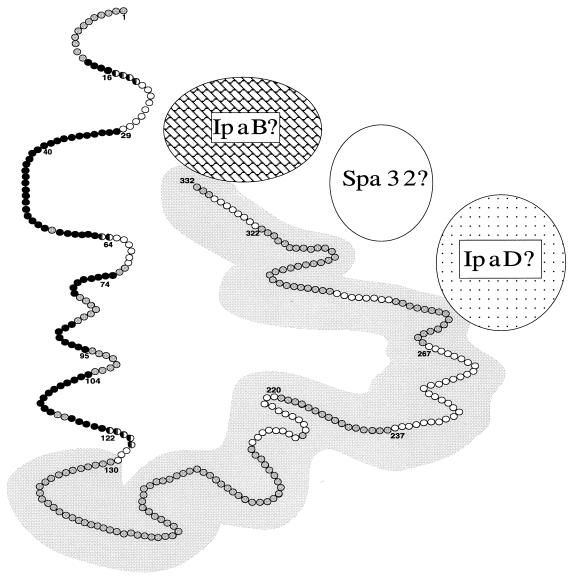
FIG. 4.
A working model of the antigenic topography of membrane-bound IpaD. This model shows epitopes of IpaD that are exposed on the surface of shigellae (black circles) and epitopes of IpaD that are not accessible to antibody (white circles). Amino acid residues found in both exposed and inaccessible epitopes are represented by a half-black, half-white circle. The grey circles represent amino acid residues not identified as part of a surface-exposed or inaccessible epitope. Amino acid residues overlying the grey background are within the region of IpaD that is homologous to the Salmonella SipD protein. Amino acid residue numbers at the beginning or end of epitopes are indicated. Based on previously published data, hypothetical associations of IpaD with membrane-associated IpaB, Spa32, and IpaD are also indicated (11, 14, 31).