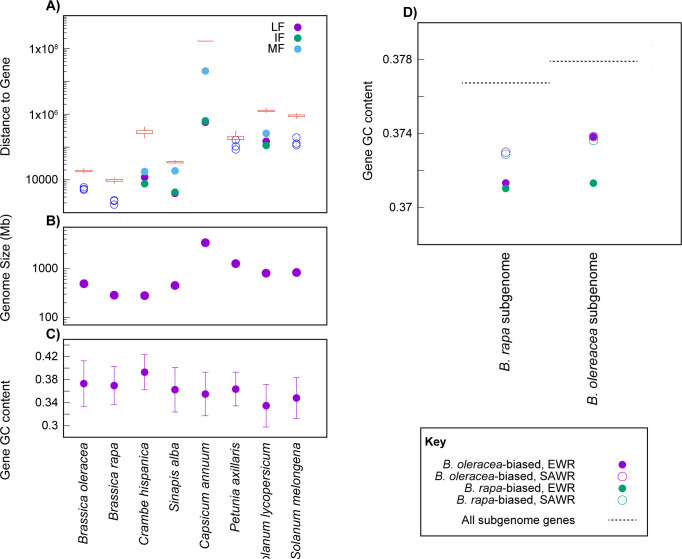
Fig 6. Mesohexaploid genomes vary several aspects of genome structure in ways relevant to the formation of further hybrids.
A) We identified tRNA genes in the intergenic region of each of eight genomes with shared mesohexaploidies and then computed the mean distance of those genes to the nearest protein coding gene for which we could identify the subgenome of origin (y-axis; see Methods). Those subgenomes vary in their level of gene preservation from most surviving genes (least fractionated, LF), through intermediate and most fractionated (IF and MF). We compared these distances to randomly distributed tRNAs, finding that in all cases, the tRNAs were closer to surviving genes than expected (P = 0.02 for P. axillaris and P<0.01 for all other genomes; note the log-scale on y). In four cases, the subgenomes differed from each other in their mean distance to the tRNA genes more than would be expected (colored points). Genomes also differ in total size (B) and in the average GC content in the genes (mean ±2 standard deviations; C). D) Finally, both European and South Asian winter rapeseed accessions of B. napus (EWR and SAWR, respectively) show lower average gene GC content for genes with highly biased expression between paralogous pairs compared to most paralogous gene pairs (Methods).