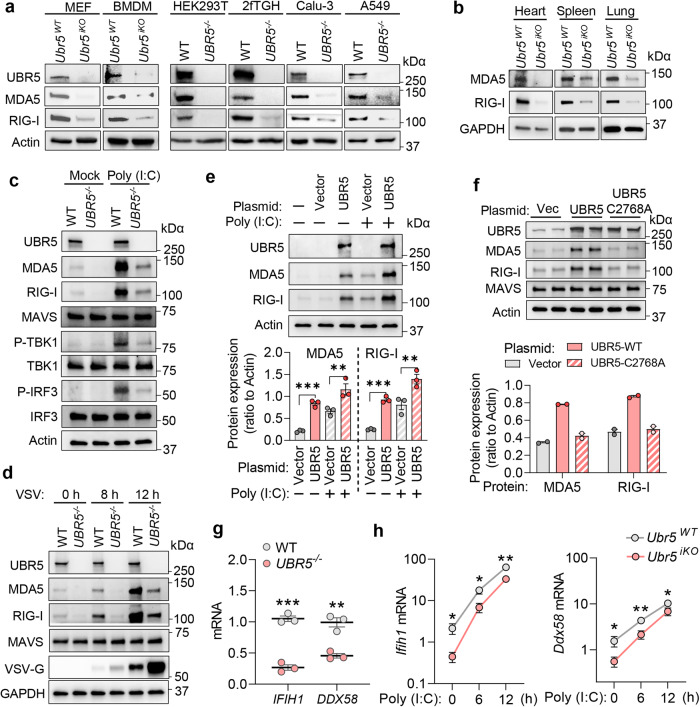
Fig. 5. UBR5 promotes RLR transcription.
The immunoblots of indicated proteins in (a) mouse primary MEFs, macrophages and various human cell lines, (b) the tissues of age- and sex-matched littermates. The immunoblots of indicated proteins in HEK293T cells (c) transfected with poly (I:C)/without (Mock) for 12 h, and (d) infected with VSV at a multiplicity of infection (MOI) of 0.5. e The immunoblots of indicated proteins in UBR5−/− HEK293T cells transfected with a UBR5 expression or vector plasmid for 24 h and then poly (I:C) (+) for 12 h. The bar chart indicates the ratios of MDA5/RIG-I band density to Actin. f The immunoblots of indicated proteins in WT HEK293T cells transfected with a vector, wild-type UBR5, or catalytic mutant C2768A plasmid. The bar chart indicates the ratios of MDA5/RIG-I band density to Actin. n = 2 biologically independent experiments. qRT-PCR quantification of the IFIH1/DDX58 (gene symbol for MDA5/RIG-I) mRNA levels in (g) HEK293T cells, and (h) MEFs transfected with poly (I:C). The data are representative of three independent experiments (a, c, d) or tissues from two mice (b) with similar results. The data shown in (e) are from one representative experiment of n = 3 biological independent experiments, mean ± S.E.M, ordinary one-way ANOVA with Dunnett’s test, ***p = 0.0008, **p = 0.0036 for MDA5; ***p = 0.0009, **p = 0.0023 for RIG-I. Multiplicity adjusted p values are presented. Data shown in g, h: mean ± S.E.M, two-tailed Student’s t test, n = 3 biologically independent experiments; ***p = 0.0004, **p = 0.0028 in g; for h: *p = 0.0493, *p = 0.0458, **p = 0.0017 in sequence for Ifih1; *p = 0.0286, **p = 0.0086, *p = 0.0472 in sequence for Ddx58. Adjusted p values are presented. Source data are provided as a Source Data file.