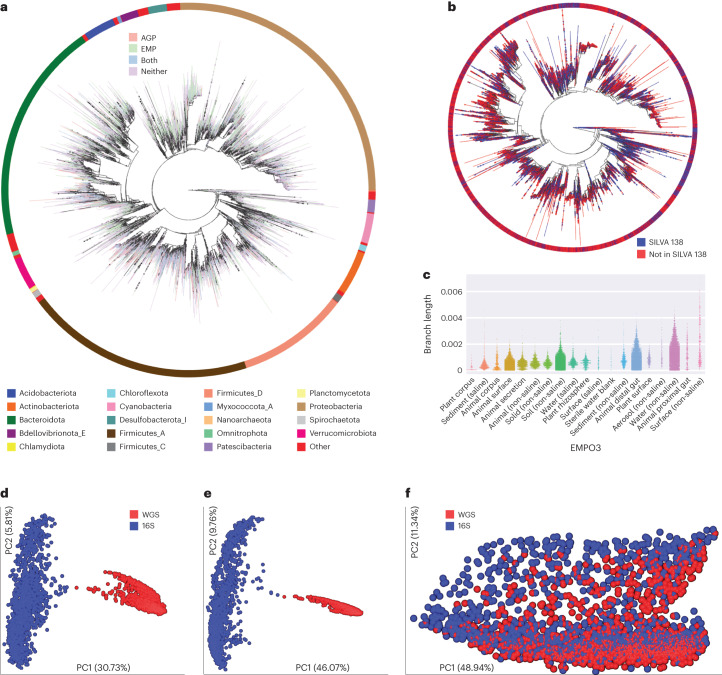
Fig. 1. Greengenes2 overview and harmonization of 16S rRNA ASVs with shotgun metagenomic data.
a, The Greengenes2 phylogeny rendered using Empress23, with ASV multifurcations collapsed; tip color indicates representation in the American Gut Project (AGP), the EMP, both or neither, with the top 20 represented phyla depicted in the outer bar. b, The same collapsed phylogeny colored by the presence or absence of the best BLAST24 hit from SILVA 138. The bar depicts the same coloring as the tips. c, EMP samples and the amount of novel branch length (normalized by the total backbone branch length) added to the tree through ASV fragment placement. Note that sample counts are not even across EMPO3 categories. d, Bray–Curtis applied to paired 16S V4 rRNA ASVs and whole-genome shotgun samples from THDMI subset of The Microsetta Initiative; PC, principal coordinate. e, Same data as d but computing Bray–Curtis on collapsed genus data. f, Same data as d and e but using weighted UniFrac at the ASV and genome identifier levels.