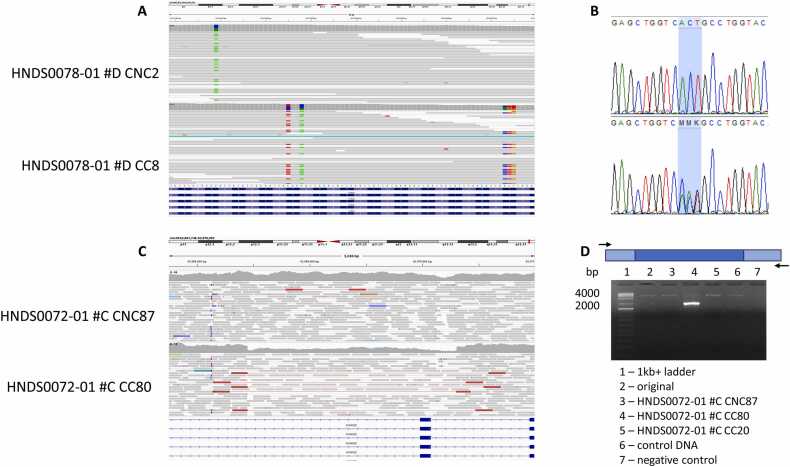
Fig. 2.
On-target 3-bp substitution detected by Off-flow in CRISPR/Cas-edited clone HNDS0078–01 #D CC8 and on-target mono-allelic deletion detected by Off-flow in CRISPR/Cas-edited clone HNDS0072-01 #C CC80. (A) IGV visualization. The colored portions of reads represent mismatched bases. Each color represents a different nucleotide, green represents A, blue represents C, red represents T and orange represents G. If all reads match the reference genome, the coverage bar is gray. If a nucleotide differs from the reference sequence in greater than 20% of quality weighted reads, the bars are colored in proportion to the read count of each base. (B) Sequence trace showing the 3 bp substitution in KCNQ2 in cell line HNDS0078–01 #D CC8. (C) IGV visualization. Deletions are displayed with a red bar. The length of the bar indicates the size of the deletion. (D) Schematic of PCR amplicons. Dark blue rectangle shows location of deletion. Arrows how the primers used for PCR (top panel). Agarose gel electrophoresis of ∼4000 bp PCR amplified fragment reveals a single band of ∼2000 bp from cell line HNDS0072–01 #C CC80 that was not present in other clones (bottom panel).