Abstract
The ascomycete Podospora anserina is a heterothallic filamentous fungus found mainly on herbivore dung. It is commonly used in laboratories as a model system, and its complete life cycle lasting eight days is well mastered in vitro. The main objective of our team is to understand better the global process of fruiting body development, named perithecia, induced normally in this species by fertilization. Three allelic mutants, named pfd3, pfd9, and pfd23 (for “promoting fruiting body development”) obtained by UV mutagenesis, were selected in view of their abilities to promote barren perithecium development without fertilization. By complete genome sequencing of pfd3 and pfd9, and mutant complementation, we identified point mutations in the mcm1 gene as responsible for spontaneous perithecium development. MCM1 proteins are MADS box transcription factors that control diverse developmental processes in plants, metazoans, and fungi. We also identified using the same methods a mutation in the VelC gene as responsible for spontaneous perithecium development in the vacua mutant. The VelC protein belongs to the velvet family of regulators involved in the control of development and secondary metabolite production. A key role of MCM1 and VelC in coordinating the development of P. anserina perithecia with gamete formation and fertilization is highlighted.
Keywords: perithecia, Podospora anserina, development, sexual reproduction, ascohymenials
1. Introduction
Ascomycota filamentous fungi differentiate complex multicellular fruiting bodies during sexual reproduction. Fruiting body formation usually requires a set of proper environmental conditions, such as light, temperature, and proper nutrients. In the ascohymenials, elaboration of most of the tissues of the fruiting body takes place after fertilization through precise developmental programs involving differentiated cells (for review [1,2]). On the contrary, in the ascoloculars, most tissues of the fruiting bodies are formed before the gametes, and hence before fertilization. The molecular mechanisms and the complete regulatory scheme leading to the production of mature fruiting bodies are not yet fully characterized despite numerous studies [2,3,4,5]. Different signal transduction pathways have been shown to control fruiting body development, and these include the MAP kinases, the STRIPAK complex, and the NADPH oxidases [1,5,6,7]. Recent comparative genomic and transcriptomic approaches have shown a large number of regulated genes during fruiting body development [8,9], but their roles remain to be determined. Although it produces fewer data than genomic methods, the classical approach of genetic analysis is a powerful tool to identify key genes involved in fruiting body development and, through a thorough phenotypic analysis, to obtain important information about their roles in the process.
The filamentous ascomycete Podospora anserina is a good model to study development of fruiting bodies (called perithecia in this species) through a direct genetic approach [2,7,10,11]. In P. anserina, the three-dimensional perithecia are issued from two types of cells. The envelope (or peridium) is derived from maternal vegetative haploid mycelium, whereas the dikaryotic zygotic tissue comes from both male and female parents and is embedded in the centrum (the tissues within the peridium that have both a maternal origin, such as the paraphyses, and a zygotic origin, i.e., the sexual lineage producing asci). Female gametangia, ascogonia, and male gametes, spermatia, are differentiated first (ascohymenial development). The ascogonium is rapidly surrounded by maternal hyphae and becomes a preformed fruiting body, the protoperithecium [2], which awaits fertilization. After fertilization, there is a coordinated growth of the maternal and zygotic tissues and perithecia begin to mature. After two days, necks and ostioles are formed and, after four days, maturation is completed with ascospore production and projection. There is thus a need to coordinate the developments of both the zygotic and maternal tissues in order to orchestrate the whole fruiting body development correctly.
In order to understand the developmental program leading to the development of perithecia in P. anserina better, mutageneses on wild-type homokaryotic strains, i.e., unable to engage in mating, have been performed to isolate mutants able to produce perithecia without prior fertilization. They have an identical phenotype to that of the long-known vacua mutant [12]. These mutants are called pfd, for “promoting fruiting body development”. We describe here the characterization of some of them that are in fact alleles of the same gene encoding the MADS box protein orthologous to the Saccharomyces cerevisiae MCM1 [13,14] and identify the gene affected in the vacua mutant as encoding the ortholog of Aspergillus nidulans VelC, a protein of the velvet family.
2. Materials and Methods
2.1. Strains and Growth Conditions
All strains of P. anserina used in this study are derived from the “S” (big S) wild-type strain that was used for sequencing of the P. anserina genome [15]. The most recent protocols for standard culture conditions, media, and genetic methods for P. anserina are described by Silar [16]. The Δmus51::phleoR mutant strain differs from the S wild-type reference strain by a single deletion of the mus-51 gene, which has increased frequency of targeted gene replacement [17]. The pks1-193 strain has a mutation in the PKS1 gene that encodes a polyketide synthase necessary for mycelium pigmentation [18]. The Δmat strain, lacking its mating type, is unable to engage in fertilization, but is fully able to provide hyphae for the building of the maternal tissues of the fruiting bodies [2,19]. These two last strains have been used in sexual crosses involving some pfd mutants and mcm1Δ. The vacua strain was purchased from ATCC (ATCC n° 42805). This mutant was obtained in the second strain of P. anserina: the s (small s) strain [12]. So, the first step was therefore to introgress the vacua mutation into the S (big S) background by ten successive crosses with the S strain as the female parent. In each successive generation, the 2:2 segregation of the vacua phenotype was observed, indicating as previously stated [12] that the phenotype was due to a single mutation.
2.2. Mutagenesis
Two UV mutageneses were made. In the first one (mutagenesis I), protoplasts (obtained as described in Silar [16]) of the S mat- strain were spread onto regeneration plates [16] at 104/plate and irradiated with UV 256 nm at a dose of 250 J/m2, which resulted in the death of 99% of the protoplasts. The plates were then incubated at 27 °C in the dark. Thalli generated two days later by surviving protoplasts were transferred onto M2 plates at nine thalli per plate and incubated for a week at 27 °C in the presence of light (equivalent to daylight). Thalli generating perithecia/protoperithecia larger than those of the wild type were selected. Among 3000 tested protoplasts, 14 were found to contain a mutation enabling the development of large fruiting bodies, although some lack a neck and ostiole.
In the second round of mutagenesis (mutagenesis II), two-day-old S mat+ thalli were fragmented in a Precess 24 (Bio-Rad, Hercules, CA, USA) (speed 4.0 for 20 s) and spread onto M2 medium. After an overnight incubation, the regenerating thalli were irradiated with UV 256 nm at a dose of 300 J/m2, protected from light overnight, and then incubated for a week at 27 °C in the presence of light. Large protoperithecia/fruiting bodies appearing on the plates were then transferred with some neighboring hyphae onto fresh M2 medium for further analysis. Thirteen additional mutants among 50 plates were recovered by this method. The resulting 27 mutants were then individually identified and numbered as pfd mutants, the term pfd meaning “Promoting Fruiting body Development” (Table 1).
Table 1.
The pfd mutants.
| Mutant | Fruiting Body Formation | Linkage | Other Phenotypes | |
|---|---|---|---|---|
| Mutagenesis I | pfd1 | Large neckless protoperithecia | Fluffy | |
| pfd3 mat- | Normal-looking perithecia when in heterokaryon with the wild type of same mating type | mat (d < 1 cM) | Flat and dark mycelium, male sterility, female sterility but small protoperithecia formed | |
| pfd4 | Large neckless protoperithecia | Spindly mycelium that becomes wild type, homozygous crosses produce empty perithecia filled with jelly | ||
| pfd6 | Large neckless protoperithecia | Fluffy | ||
| pfd7 | Large neckless protoperithecia | Fluffy | ||
| pfd8 | Large neckless protoperithecia | None | ||
| pfd9 mat- | Normal-looking perithecia and large neckless protoperithecia | mat (d < 1 cM) | None | |
| pfd10 | Large protoperithecia some with neck initials | Slow and fluffy growth, homozygous crosses are partially sterile | ||
| pfd11 | Large neckless protoperithecia | Pink mycelium, homozygous crosses are sterile | ||
| pfd12 | Large neckless protoperithecia | Pink mycelium, homozygous crosses are sterile | ||
| pfd13 | Few normal-looking perithecia and large neckless protoperithecia | Homozygous crosses are sterile | ||
| pfd15 | Large neckless protoperithecia | None | ||
| pfd16 | Large neckless protoperithecia | Pink mycelium, homozygous crosses are partially sterile | ||
| pfd17 | Few normal-looking perithecia and large neckless protoperithecia | Fluffy, perithecia with abnormally shaped neck in homozygous crosses | ||
| Mutagenesis II | pdf18 | Large neckless protoperithecia | None | |
| pfd19 | Few normal-looking perithecia and large neckless protoperithecia | Fluffy, perithecia with abnormally shaped neck in homozygous crosses | ||
| pfd20 | Large neckless protoperithecia | Fluffy | ||
| pfd21 | Large neckless protoperithecia | None | ||
| pfd22 | Large neckless protoperithecia | None | ||
| pfd23 mat+ | Normal-looking perithecia and large neckless protoperithecia | mat (d < 1 cM) | Dark and fluffy mycelium | |
| pfd24 | Large neckless protoperithecia | None | ||
| pfd25 | Large neckless protoperithecia | None | ||
| pfd26 | Large neckless protoperithecia | None | ||
| pfd28 | Large neckless protoperithecia | Pink mycelium | ||
| pfd29 | Large neckless protoperithecia | None | ||
| pfd30 | Small perithecia with large necks | Slow-growing mycelium | ||
| pfd31 | Few normal-looking perithecia and large neckless protoperithecia | None |
All selected mutants were crossed to the wild type, and the pfd phenotype was shown to segregate as due to a single mutation. For some mutants, additional phenotypes co-segregated with the pfd phenotype, indicative of pleiotropy of the mutations. In this work, we will focus on the characterization of three allelic pfd mutants: pfd3, pfd9, and pfd23 (see results).
2.3. Identification of Genes Mutated in pfd3, pfd9, and vacua Mutants
The Illumina sequencing work benefited from the facilities and expertise of the I2BC high throughput sequencing platform (formerly IMAGIF, https://www.france-genomique.org/plateformes-et-equipements/plateforme-de-sequencage-haut-debit-de-li2bc/, accessed on 4 December 2023). Custom-made libraries had 300 bp inserts and sequencing was 76-bp paired-end. Coverage was 85-fold. Sequence reads were mapped onto the reference genome of the S mat+ strain [15] and potential mutations were detected using the galaxy server (https://usegalaxy.org, accessed on 4 December 2023). Briefly, fastq reads were mapped onto the S reference genome with Bowtie2 version 2.0.5 [20]. Polymorphisms with the reference sequence were identified with the samtools mpileup [21] and filtered with VarScan mpileup version 2.4.3 [22].
2.4. Characterization of the Mutation in pfd23 Mutants
To identify mutations in the pfd23 mutants, the complete CDS of mcm1 (Pa_1_19280) was amplified from mutant genomic DNA with primers mcm1-1F/mcm1-4R (Supporting Information Table S1), using a high-fidelity polymerase, and then sequenced.
2.5. Phylogenetic Analysis
Fungal genes orthologous and paralogous to vacua were searched for by BLAST 2.8.1 [23] in GenBanK and Mycocosm in February 2019 [24] using the default parameters, with the vacua protein as the query. Alignment was made with MAFFT V7.419 [25] and manually refined with Jalview 2.10.2 [26]. This alignment was used to construct a phylogenetic tree using the maximum likelihood method with 100 bootstrap replicates (PhyML software V2.4.4 using the default parameters) [27] and transferred to the iTOL server V5.1 for visualization [28].
2.6. Deletion of mcm1 and ste12 Genes
The deletion cassette of mcm1 (Pa_1_19280) was constructed, as previously described [29]. The construction of the ste12 (Pa_7_1730) deletion cassette was based on the AscI/MluI method [30], modified as described by Coppin et al. [31]. A hygromycin resistance cassette was used for both. Protoplasts from the mus51Δ::phleoR and mus51Δ::su8 were transformed by the deletion cassettes of mcm1 and ste12, respectively. Transformants were selected on medium containing 75 μg/mL hygromycin. Putative deleted mutants were first selected by PCR. The PCR amplified specific junctions to the replaced locus, using two primers that annealed at one end of the hygromycin resistance gene and upstream of the proximal flank used in the deletion cassette (Supporting Information Table S1). Homologous recombination of the deletion cassette allowed amplification of a predictable fragment on each side of the selectable resistance gene. Two or three candidate transformants were genetically purified by crossing them to the wild-type strain to eliminate potential untransformed nuclei and to segregate out the mus-51Δ mutation. Several mat+ and mat- strains containing the deletion but lacking mus-51Δ were selected from each progeny and subjected to Southern blot analysis for final validation (Supporting Information Figure S1). Purified transformants verified by Southern blot were chosen as stock deletion mutants for subsequent studies. For mcm1, transformation experiments were performed in both mating types. The double mutants mcm1Δste12Δ, in both mating types, were constructed by genetic crosses, and relevant genotypes were confirmed by PCR.
2.7. Complementation of the pfd3, pfd9, pfd23, mcm1Δ, and vacua Mutants
To ensure that the phenotypes observed for mcm1 mutants were actually due to inactivation of the relevant gene, a complementation procedure was performed by ectopic insertion of the corresponding wild-type allele under control of its native promoter. A 1952 bp-DNA fragment carrying the wild-type mcm1 CDS (Pa_1_19280) and its 5′ and 3′ flanking regions was amplified with primers mcm1-1F/mcm1-4R (Supporting Information Table S1), using a high-fidelity polymerase. The DNA fragment was cloned at the Eco RV site of the pBC-Phleo plasmid [32] to yield plasmid pBC-mcm1. Protoplasts of the mcm1Δ mat+ mutant strain were transformed with this plasmid. Thirty-one transformants were selected on medium containing 10 μg/mL phleomycin. They were crossed to the mcm1Δ mat- strain. Eighteen transformants recovered a wild-type phenotype. Lack of fertility in the thirteen remaining transformants was likely due to either DNA rearrangements during integration and/or lack of expression at the insertion point. Three fertile transformants were selected and crossed with the wild type. Progeny was recovered and analyzed to identify F1 descendant strains carrying a mcm1 wild-type transgene associated with the mcm1 mutant allele or mcm1 wild-type allele. These strains were then used to verify complementation for all mcm1 mutants by crosses.
To complement the vacua mutation, the wild-type Pa_7_6330 allele was amplified by PCR using primers vacuAfor and vacuArev (Supporting Information Table S1) on genomic DNA from the S strain. The vacua mutant was then co-transformed using 10 µg of the amplified product and 2 µg of pBC-hygro. Seventeen hygromycin B-resistant transformants were obtained, among which one complemented the vacua phenotype, i.e., did not spontaneously produce large perithecia.
2.8. Generation of the mcm1 Over-Expression Strains
To overexpress mcm1, three DNA fragments were amplified with Thermo Scientific Phusion High-Fidelity DNA Polymerase: a 750 bp DNA fragment carrying the promoter of the AS4 gene using primers pBC-rev/AS4_spod1R, a 941 bp-DNA fragment carrying the CDS of mcm1 with primers Surexmcm1-For/Surexmcm1-Rev, and a 216 pb-DNA fragment carrying the ribP2 terminator with primers TT_mcm1/pBC-For. All the primers are summarized in Table S1. These three fragments were fused by PCR with primers pBC-rev/pBC-For. The DNA fragment was cloned into the pBC-Geneticin plasmid [33], previously digested by EcoRV to yield pBC-surexmcm1. This plasmid was transformed into the protoplasts of the mcm1Δ mat+ mutant strain. Twenty-one transformants were selected on medium containing 250 μg/mL geneticin. They were crossed to the mcm1Δ mat- mutant. Seven transformants recovered a wild-type phenotype. Progeny of three transformants crossed with the wild-type strain were recovered, and F1 descendants carrying an overexpressed mcm1 transgene associated with the mcm1 wild-type allele were selected for analysis. The resulting strain was named surexmcm1.
2.9. Construction of the mcm1-GFP Strains
To express the mcm1-GFP gene, three DNA fragments were amplified with Thermo Scientific Phusion High-Fidelity DNA Polymerase: a 741 bp DNA fragment carrying the promoter of the AS4 gene using primers AS4-1F/AS4_mcm1R, a 896 bp-DNA fragment carrying the CDS of mcm1 with primers Mcm1_AS4_F/Mcm1_fluo_R, and a 956 pb-DNA fragment carrying the CDS of GFP associated with the ribP2 terminator with primers Fluo_97F/TT_216R. All the primers are summarized in Table S1. These three fragments were fused by PCR with primers AS4-1F/TT_216R. The DNA fragment was cloned into the pBC-Geneticin plasmid [33], previously digested by EcoRV to yield the pBC-mcm1_GFP plasmid. This plasmid was transformed into the protoplasts of the mcm1Δ mat+ mutant strain. Seven transformants were selected on medium containing 250 μg/mL geneticin. They were crossed to the mcm1Δ mat- mutant. Three transformants recovered a wild-type phenotype, demonstrating that the addition of eGFP did not modify the activity of mcm1. Progeny of these transformants crossed with the wild-type strain were recovered and a dozen F1 descendants carrying the mcm1-GFP transgene associated with the mcm1 allele were selected for analysis. The strains were crossed with the wild type, and mat+ and mat- strains expressing MCM1-GFP were selected in the progeny.
Strains expressing both MCM1-GFP and a histone H1-mCherry fusion protein were recovered in the progeny of a cross between a strain carrying MCM1-GFP and a strain carrying the histone H1-mCherry fusion protein previously constructed [34].
2.10. Growth and Development
To determine the role of MCM1 in fungal physiology, the mutated and deleted mutants and the wild-type strain were incubated separately on M2 medium at 27 °C. Colony morphology, pigmentation, perithecium formation, ascospore production, ascospore dispersal, and germination were observed during their vegetative growth and their sexual cycle.
Perithecium content was fixed in 7.4% paraformaldehyde and analyzed, as in Coppin et al. [31]. Microscopic observations with eGFP and mCherry were made on mycelia grown on agar. Pictures were taken with a Leica microscope (Bannockburn, IL, USA) DMIRE2; LED Lumencor (optoprim): eGFP (485/20) at the ImagoSeine Plateform of the Institut Jacques Monod (https://www.ijm.fr/technological-resources-and-facilities/platforms/imagoseine/?lang=en, accessed on 4 December 2023) or with a Nikon eclipse Ci microscope. Pictures were analyzed with ImageJ (Wayne Rasband, NIH, http://imagej.nih.gov/ij, accessed on 4 December 2023).
Microscopic observations of perithecia were made of mycelia grown on agar and images were captured with a home-made microscope, combining a camera (monochromatic 1/3” CMOS sensor with a resolution of 1280 × 960 pixels) and a telecentric objective (4× magnification and 65 mm focal length). The precision of the microscope was measured to be 1.10 micro-m/pixel. Enlightenment was achieved using the beam from a white LED light passing through the sample.
Perithecia enumeration and area estimation were performed using Aphelion Imaging Software V4.4.0 (ADCIS SA and Amerinex Applied Imaging), the different steps of which are detailed in Supporting Information Figure S2. Anastomoses were analyzed, as in Chan Ho Tong et al. 2014 and in Brun et al. 2009 [33,34].
2.11. Sensitivity to Various Stresses
Sensitivity of the mutated and deleted mutants was tested on sorbitol (0.55 M), KCl (0.25 M), NaCl (0.25 M), Congo red (0.7 mM), calcofluor (50 µM), and methylglyoxal (2.5 mM).
Sensitivity to antifungals was measured with Etest® gradient strips (bioMérieux, Marcy l’Etoile, France) with fluconazole (0.016–256 µg/mL), amphotericin B, and itraconazole (0.002–32 µg/mL). The Etest® assay was performed in accordance with the manufacturer’s instructions. Plates were inoculated with mycelia fragmented in a Precess 24 (BioRad, Hercules, CA, USA) onto the surface using glass beads, and the antifungal agent strips were placed on the plates. The Etest® minimum inhibitory concentration (MIC) was recorded as the lowest concentration of an antifungal drug for which the elliptical zone of growth inhibition intersected the Etest strip. Microcolonies within the ellipse were ignored. The Etest® MIC was recorded after incubation for 4 days.
3. Results
3.1. Mutant Recovery
“Promoting fruiting body development” (pfd) mutants were obtained via two UV mutagenesis rounds (and were selected for their ability to promote some perithecium development without fertilization). Twenty-seven mutants were selected. They were crossed to the wild-type strain and the descendants were analyzed. The mutant phenotypes with regard to fruiting body production and vegetative growth are summarized in Table 1. Where possible, the purified pfd mutants were crossed with themselves and, in all cases, they were crossed to the wild-type strain of the opposite mating type. The pfd mutants were affected at various stages of their vegetative and sexual development. Some strains had an altered mycelium, either in its aspect, as fluffy or flat, or in its color, as dark- or pink-pigmented, when compared with the wild-type strain (Table 1). Most pfd mutants produced only large neckless protoperithecia. Yet, few mutants produced additional normal-looking perithecia that turned out to be barren.
In three mutants, pfd3, pfd9, and pfd23, the mutations were shown to be closely linked to the mating type (d < 1 cM) and could not be dissociated from it. Therefore, pfd3 and pfd9 were only available as mat-, and pfd23 as mat+. This observation led us to hypothesize that pfd3, pfd9, and pfd23 could be affected in the same gene and they were chosen for further analyses. The pfd9 and pfd23 mutants presented many normal-looking and few large neckless barren perithecia. The third one, pfd3, presented a peculiar phenotype. Indeed, it was male- and female-sterile, yet it produced barren perithecia when present in heterokaryons with wild-type nuclei of the same mating type.
3.2. Mutations
The pfd3 and pfd9 mutations were identified by direct genome sequencing. In each case, the mutant strain selected for sequencing originated from six generations of successive crosses between the mutant strain and the wild type. In this way, only 2.5% of the genome of the selected mutant strain came from the original mutant, then limiting the possibility of having mutations unrelated to the mutant phenotype. Point mutations were found for both mutants in the Pa_1_19280 CDS, when compared with the wild-type strain. In pfd3, one nucleotide substitution (C to T, position 22) and one deletion (position 24) were found at the 8th codon, resulting in a stop codon at the beginning of the CDS (Figure 1). In pfd9, a T was mutated into a C at position 298, resulting in a substitution from a leucine to a phenylalanine at codon 99 (Figure 1). By sequencing the Pa_1_19280 CDS in pfd23, one deleted nucleotide (position 510) was found, resulting in a frameshift at codon 170 (Figure 1). Overall, this confirmed that the three mutants were likely affected in the same gene.
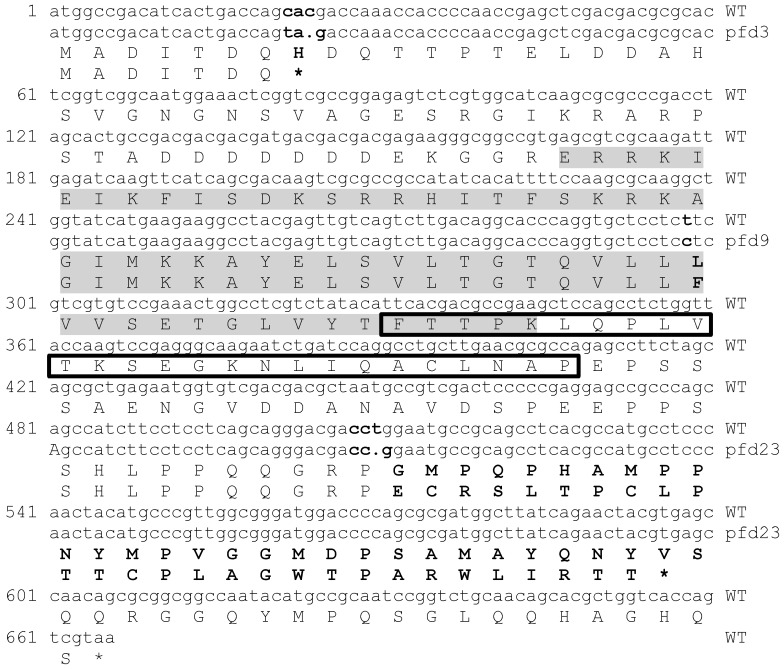
Figure 1.
Pfd mutations in the Pa_1_19280 gene and their outcome in the MCM1 protein sequence. The complete CDS of the mcm1 wild-type (WT) strain is made up of 666 nucleotides (222 codons, 221 aa). Modifications in the genetic sequences of the three mutant strains and in the resulting proteins, when compared with the WT sequence, are shown in bold type. The deletion of a nucleotide is represented by a dot in the nucleotide sequence of the concerned mutants. The remarkable signature sequences for the MCM1 proteins, i.e., the MADS box and SAM domain, are marked in the figure by a shaded area and a boxed area, respectively. * are translation stops.
Blast analysis revealed that Pa_1_19280 likely encodes the P. anserina orthologue of the MCM1 protein, which displays identity values of 82%, 84%, and 80% with the S. cerevisiae, Sordaria macrospora and Magnaporthe oryzae proteins, respectively. Blast analysis of P. anserina MCM1 also allowed the localization of two highly conserved domains, the MADS box and the SAM domain, which are conserved in canonical MCM1 (Figure 1).
The pfd9 mutation occurs in the highly conserved signature domain of the MADS box, whereas the pfd23 mutation truncates the C-terminus, which could destabilize the protein. The pfd3 mutation leads to a very short protein of 7aa and the next start codon that could be used to initiate translation is in the middle of the MADS box; pfd3 is therefore likely a null allele.
3.3. Deletion of mcm1 and Complementation
In order to ascertain whether pfd3 is a null allele and also to obtain a null allele of the opposite mating type, strains deleted in mcm1 were constructed in both mating type by targeted gene deletion. Both deleted strains had indeed the same phenotype as pfd3, i.e., they were sterile and spontaneously produced barren perithecia, but only in heterokaryons with the wild-type nuclei of the same mating type.
To verify that the deletion of mcm1 is responsible for the different phenotypes observed in the mcm1∆ strain, a wild-type allele of mcm1 was reintroduced in the deleted strains. The plasmid pBC-mcm1 carrying the wild-type mcm1 gene as well as a phleomycin resistance cassette was thus transformed into the mcm1Δ mat+ strain. Among 31 phleomycin-resistant transformants, 18 had a wild-type phenotype: perithecia on the thalli of the complemented strains were recovered in crosses with mcm1Δ mat- and no spontaneous perithecium was observed in heterokaryotic crosses with S mat+. However, among the 18 complemented transformants, 9 of them were partially complemented, as their perithecia were barren in heterokaryotic crosses with the mcm1∆ mat- strain. It is therefore possible that the location of the mcm1 gene insertion into the genome may affect the expression of the transgene and lead to a total or partial complementation. Three fully complemented transformants were purified by crosses with the S mat- strain. In subsequent crosses, complemented strains mcm1Δ in both mating types were obtained. Crosses were also made with the different pfd strains, in order to verify if the wild-type allele was able to complement the different mutations. It did so for the three mutants, confirming that the mutations carried in mcm1 by pfd3, pfd9, and pfd23 were actually responsible for the pfd phenotype.
3.4. Sexual Development in the pfd Mutants and in mcm1Δ
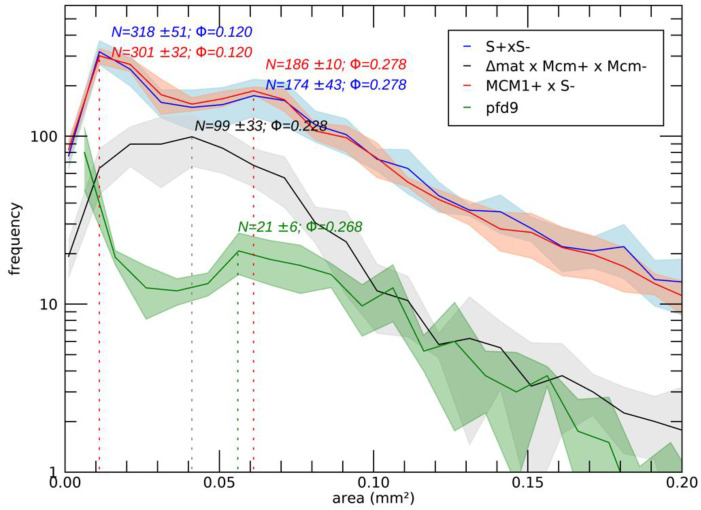
In order to study the role of MCM1 in the development cycle of P. anserina, the production of perithecia, as well as their structure and their content, was carefully observed, as it is classically performed ([16]; Figure 2). Firstly, production of perithecia by the pfd mutants was tested on G medium (medium inducing ascospore germination, see [16]), in the dark and at 30 °C. In these three conditions, the wild type did not produce perithecia, nor did the pfd mutants, including pfd3, even when associated with wild-type hyphae of the same mating type. Then, in a more original way, for each experiment, the number and the size of perithecia were carefully measured with the Aphelion software, able to detect, count, and measure small objects, such as perithecia on a Petri dish, on four independent thalli after 7 and 10 days of culture. This kind of thallus cartography allowed us to quantify the production of perithecia in the wild-type strain and in various mutants in different contexts, and allowed us to complete the observations made microscopically usefully. The details of the procedure and all the relevant measurements performed are shown in Supporting Information Figure S2. Similar results were obtained after 7 and 10 days of culture, showing that smaller perithecia sizes could not be attributed to a sexual cycle longer than the time required to obtain perithecia in a wild-type cross (7 days). Figure 3 shows the number of perithecia (frequency) as a function of the measured area (in mm2) after 10 days of culture using for each experiment with the four thalli. Typically, in the wild-type cross, we can clearly see two peaks, corresponding to two populations of fruiting bodies: (i) protoperithecia that have not matured (average number = 318 ± 51, average area ~0.001 mm2) and (ii) mature perithecia (average number = 174 ± 43, average area ~0.06 mm2). Note that the average area values obtained in this case for perithecia from a wild-type cross are consistent with those measured microscopically (Figure 2B).
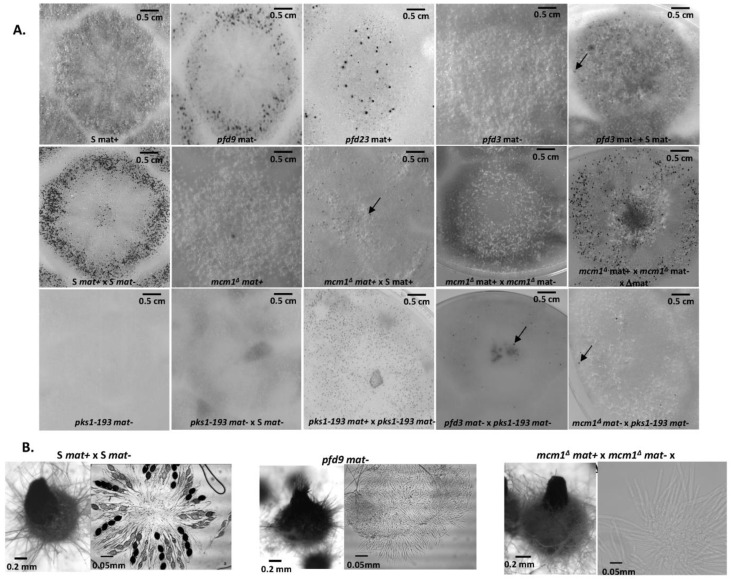
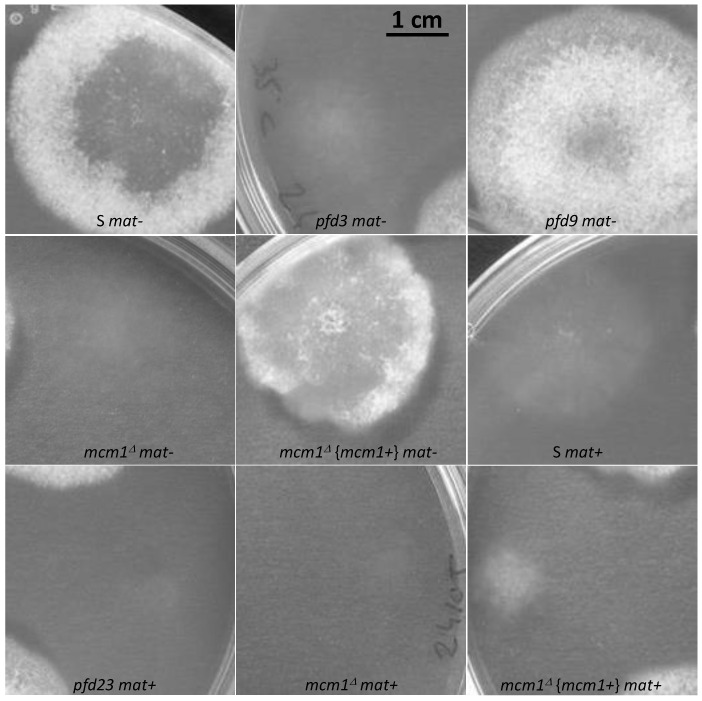
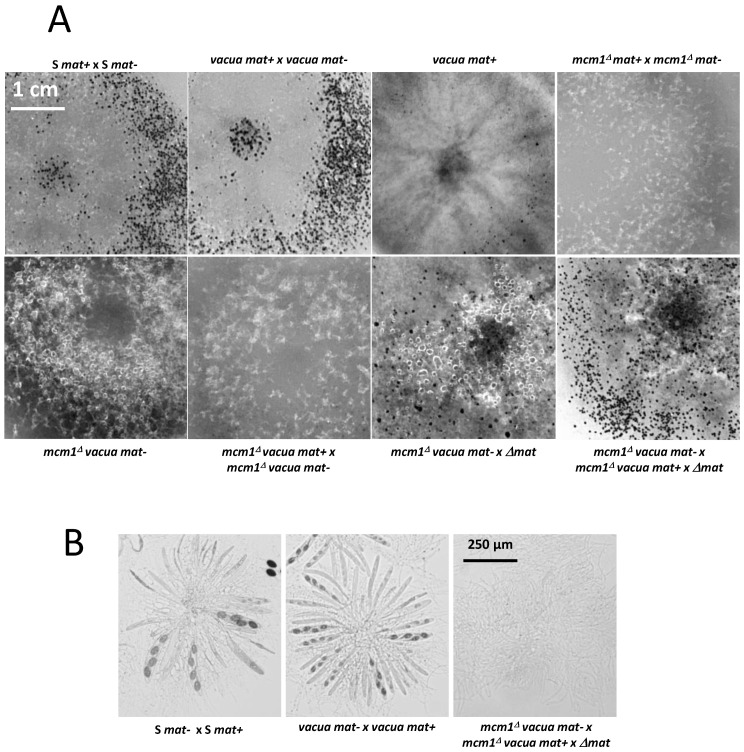
Figure 2.
Spontaneous or fertilized perithecia (heterokaryotic crosses) in pfd mutants and mcm1Δ. (A). The pictures were taken 7 days after fertilization, at which time fully mature perithecia expel ascospores. Perithecia are visible as small dots in the close-up view. Arrow indicates pfd3 and mcm1Δ spontaneous perithecia. (B). Microscopic observations of perithecia and typical rosette of asci observed in S fertilized perithecia and paraphyses observed in spontaneous perithecia after 7 days of growth in pfd9 and in mcm1Δ crosses in the ∆mat strain context. The pfd9 mutant presents various sizes of spontaneous perithecia, some of which display sizes similar to wild-type fertilized perithecia, as shown here.
Figure 3.
Number and variation in the size of fertilized perithecia after 10 days of growth (with N the average number of perithecia and Φ their equivalent diameter in mm). Perithecia metrics were extracted using Aphelion Imaging Software. For each strain, four independent replicates were performed. For details, see Supporting Information Figure S2.
The pfd9 and pfd23 mutants were able to make spontaneous perithecia on their own (Figure 2A). As expected, since there was no fertilization, all spontaneous perithecia observed in mutant strains did not contain ascospores. Even if the size of these structures remains small, there is formation of true necks and tuft of hairs, which are characteristics of P. anserina perithecia [12]. These structures were filled with paraphyses (Figure 2B). The mycelium of pfd9 and pfd23 presented with the same pigmentation as the wild-type strain, with few aerial filaments for pfd9 and a mycelium that appeared more aerial for pfd23. Neither of them presented with male or female sterility. In both cases, we counted a rather small number of perithecia (average number = 21 +/−6 for pfd9, Figure 3) but a rather wide distribution of surfaces around an average area of ~0.055 mm2 for pfd9 (Figure 3), which showed that we had a population of spontaneous perithecia in a lower number and smaller size than mature perithecia but clearly bigger than the protoperithecia measured in a wild-type cross.
As previously stated, the pfd3 mutant was found to be peculiar. Indeed, in the homokaryotic progeny of a cross between pfd3 and the wild type, we could not find any thalli forming perithecia, while the original mutant formed repetitively wild-type-looking perithecia. In fact, the pfd3 mutant was only able to make spontaneous perithecia in heterokaryotic mycelium with wild-type nuclei of the same mating type (Figure 2A), and the original mutant was likely a heterokaryon carrying wild-type and mutant nuclei. The pfd3 strain pigmented quickly and its mycelium had few airborne filaments compared with the wild-type strain. Similar phenotypes were observed for both mcm1Δ strains.
Spreading experiments involving pfd3 x S or mcm1Δ x S were carried out and showed male and female sterility for the mutant strains. Indeed, no perithecium was observed on the mycelium of pfd3 or mcm1Δ strains nor on the mycelium of the wild-type strain. No male (spermatium) or female (ascogonium) gametes were observed on the mutant thallus, probably indicating that these mutants were unable to differentiate these structures. Perithecia were only observed at the confrontation between the two strains. They were probably obtained by direct fertilization of ascogonia from the wild-type strain with mycelial filaments of the mutant strain. Another explanation was that heterokaryons may have been formed and complementation occurred where both strains met.
Then, the particularity of the pfd3 mutant is that it needs wild-type nuclei of the same sexual type to make spontaneous perithecia. To understand better whether these perithecia originated from the mycelium of the mutant or that of the wild strain, a heterokaryon was constructed with pfd3 and the pks1-193 strain of the same mating type. This strain has a mutation in the PKS1 gene that encodes a polyketide synthase necessary for mycelium pigmentation [18]. Perithecia from the mycelium of this strain are pink, while perithecia from a wild-type mycelium are dark green and pigmentation is cell-autonomous. We observed that pfd3 mat- X pks1-193 mat- heterokaryons were able to produce spontaneous barren perithecia that were fully pigmented dark green. Thus, we can conclude that these perithecia were made up solely of hyphae coming from the pfd3 mutant, but were produced only if the mycelium contained a functional MCM1.
The mcm1Δ mutant did not produce spontaneous perithecia when cultured alone (Figure 2A). As with the pfd3 mutant, the mcm1Δ nuclei must be in heterokaryons with wild-type ones of the same sexual type to allow spontaneous perithecia production. In heterokaryons with the pks1-193 strain, the spontaneous perithecia observed were dark green, indicating that maternal tissue was provided by mcm1Δ hyphae and that wild-type nuclei were necessary for the formation of these unfertilized perithecia (Figure 2A). As with the three pfd mutants, the spontaneous perithecia from the mcm1Δ strain did not contain spores and were filled with paraphyses. In spreading crosses with a wild-type strain of the complementary mating type, they also displayed male and female sterility, with a very low production of gametes, spermatia, and ascogonia. Finally, mcm1Δ mat+ X mcm1Δ mat- crosses were completely sterile. In order to understand better at which stage of sexual development the mcm1Δ strain is affected, we measured fertility in the mcm1Δ mat+ X mcm1Δ mat- X Δmat trikaryon. This strain is unable to engage in fertilization, but is fully able to provide hyphae for the building of the maternal tissues of the fruiting bodies [2,35]. Here, a partial rescue in the trikaryon was observed, indicating that mcm1 was required in the mycelium and/or the maternal tissues of the fruiting bodies for the formation of the perithecium envelope (Figure 2A). However, ascospore production was not improved in the trikaryon, showing that mcm1Δ mat+ X mcm1Δ mat- zygotic tissue was also defective. Finally, we showed that heterokaryotic crosses mcm1Δ mat+ x S mat- (Figure 3) and mcm1Δ mat- x S mat+ behave as a wild-type cross in terms of number and size of protoperithecia and perithecia, meaning that, indeed, mcm1Δ shows normal sexual reproduction when crossed to the wild-type strain of the opposite mating type. Moreover, in the trikaryon mcm1Δ mat+ X mcm1Δ mat- X Δmat, we observed a single peak, quite large (average number = 99 ± 33, average area ~0.04 mm2), corresponding to a partial rescue in the trikaryon, with a population of perithecia in a lower number and smaller size than in a wild-type cross (Figure 3).
3.5. Sensitivity to Various Stresses
Because the mcm1 gene was previously shown to be involved in cell wall and osmotic stresses in S. cerevisiae [36], we evaluated resistance/sensitivity of the mutants to these various stresses as well as to oxidative and temperature stresses. Stress tests were carried out on the pfd and mcm1Δ mutant strains, the complemented strain and the surexmcm1 strain, in which mcm1 CDS is under the control of the strong and constitutive promoter of the AS4 gene encoding the translation elongation factor EF1α. Each experiment was performed in triplicate. As the surexmcm1 strain behaved throughout the experiments as the wild-type strain, this strain was not further analyzed. All tested strains behaved like the wild-type strain under osmotic (sorbitol, NaCl and KCl) and oxidative (methylglyoxal) stress conditions.
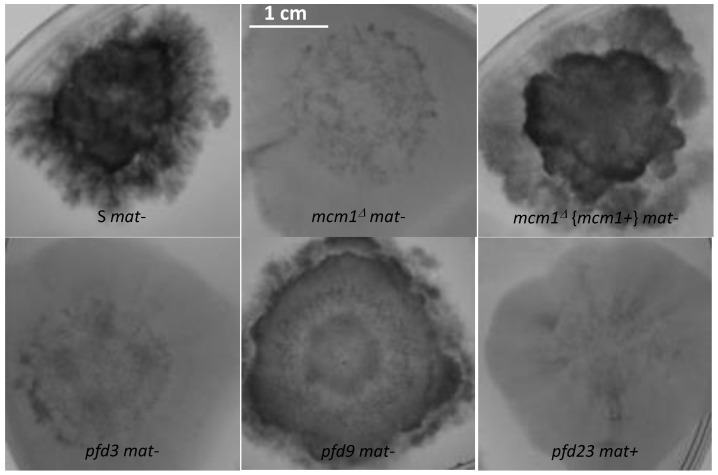
Calcofluor, which binds to chitin, and Congo red, which inhibits glucan synthases of the cell wall, had similar effects on the cell wall of the mutants (Figure 4). Briefly, the complemented strain behaved like the S wild-type strain, with a dark-pigmented mycelium and severely impaired growth. By contrast, the pfd3, pfd23, and mcm1Δ mutants showed normal growth that did not appear to be affected by the presence of both compounds. To a lesser extent, the pfd9 mutant seemed to have an intermediate phenotype with little growth impact, only exhibiting a dark-pigmented mycelium.
Figure 4.
Effect of calcofluor (50 μM) on mycelial growth. Mutant and wild-type strains were observed after 3 days.
Temperatures below (18 °C) and above (35 °C) the optimal growth temperature (27 °C) for P. anserina were tested. At 18 °C, all strains behaved in the same way as the wild-type strain. At 35 °C (Figure 5), the usual heat resistance of the S mat- strain was found when compared with the S mat+ strain, as previously described [12]. This phenotype of heat resistance was found in pfd9 mat- but not in pfd3 mat- nor in mcm1Δ mat-.
Figure 5.
Effect of high temperature (35 °C) on mycelium growth. Pictures were taken after 3 days of growth.
3.6. Effects of Antifungals
As MCM1 was previously described to be involved in azole antifungal resistance in Candida albicans [37], resistance tests to two azole antifungals (fluconazole and itraconazole) and a polyene (amphotericin B) were evaluated with the different strains using the Etest strip technique.
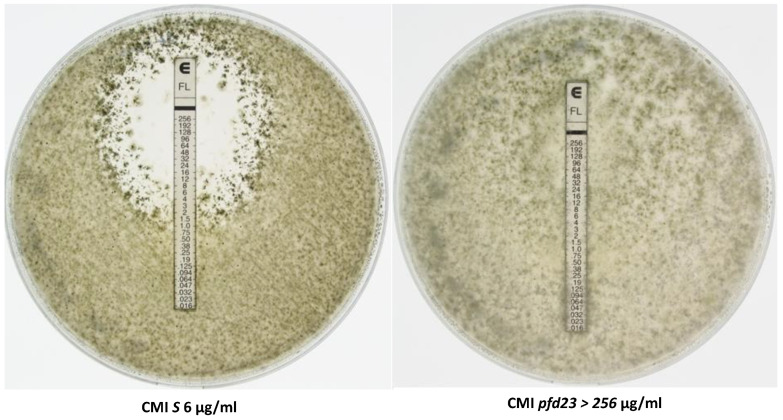
Antifungal tests with fluconazole are shown in Figure 6 and in Table 2. The S strain displayed a MIC of 6 µg/µL. The pfd3, pfd9, and mcm1Δ mutants showed a moderate resistance, with a MIC of 12 µg/µL for pfd3 and 16 µg/µL for pfd9 and mcm1Δ, while the pfd23 strain had a high resistance (MIC > 256 µg/µL). Tests with itraconazole yielded similar results (Table 2). Tests with amphotericin B showed that the pfd3 and mcm1Δ mutant strains behaved as the wild-type strain. The pfd9 and pfd23 mutants had a moderate resistance to amphotericin B, with a MIC of 0.19 µg/µL and 0.25 µg/µL, respectively.
Figure 6.
Fluconazole Etest. Each experiment was performed in triplicate. One hundred μL of fragmented mycelia from a 5 × 5 mm standardized plug was spread on the Petri dish. Pictures were taken after two days of growth.
Table 2.
Antifungal Etests.
| Strains | MIC Fluconazole | MIC Itraconazole | MIC Amphotericin B |
|---|---|---|---|
| S | 6 µg/µL | 0.5 µg/µL | 0.047 µg/µL |
| mcm1∆ | 16 µg/µL | 2 µg/µL | 0.032 µg/µL |
| pfd3 | 12 µg/µL | 1.5 µg/µL | 0.032 µg/µL |
| pfd9 | 16 µg/µL | 1.0 µg/µL | 0.19 µg/µL |
| pfd23 | >256 µg/µL | >32 µg/µL | 0.25 µg/µL |
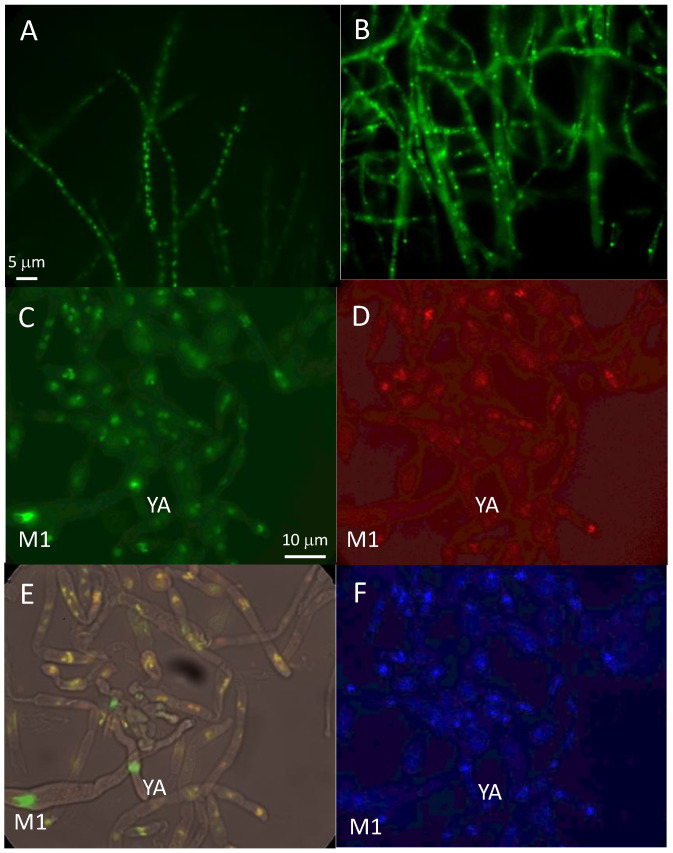
3.7. Localization of MCM1
After transformation of the mcm1∆ mat+ strain to verify that the MCM1-GFP fusion protein was functional, we observed a punctuated marking, probably corresponding to nuclear localization, in the transformants that regained a wild-type phenotype (Figure 7A,B). To confirm this, a cross with the strain carrying the MCM1-GFP with a strain possessing the histone H1-mCherry fusion protein, previously constructed [34], was realized. Images taken by epifluorescence microscopy are shown in Figure 7C–F. Punctuated fluorescence was observed inside the perithecia. Co-location experiments with H1-mCherry and DAPI staining show that distribution of the MCM1 protein was compatible with a nuclear location.
Figure 7.
Localization of MCM1. A-B PaMCM1-GFP in vegetative phase of the mcm1∆ mat+ strain, (A) exponential phase, (B) stationary phase. (C–F) PaMCM1-GFP during sexual development, C PaMCM1-GFP; D- H1-mCherry; E- merge; F DAPI. YA: young ascus, M1: ascus in prophase of meiosis 1.
3.8. Interaction with vacua
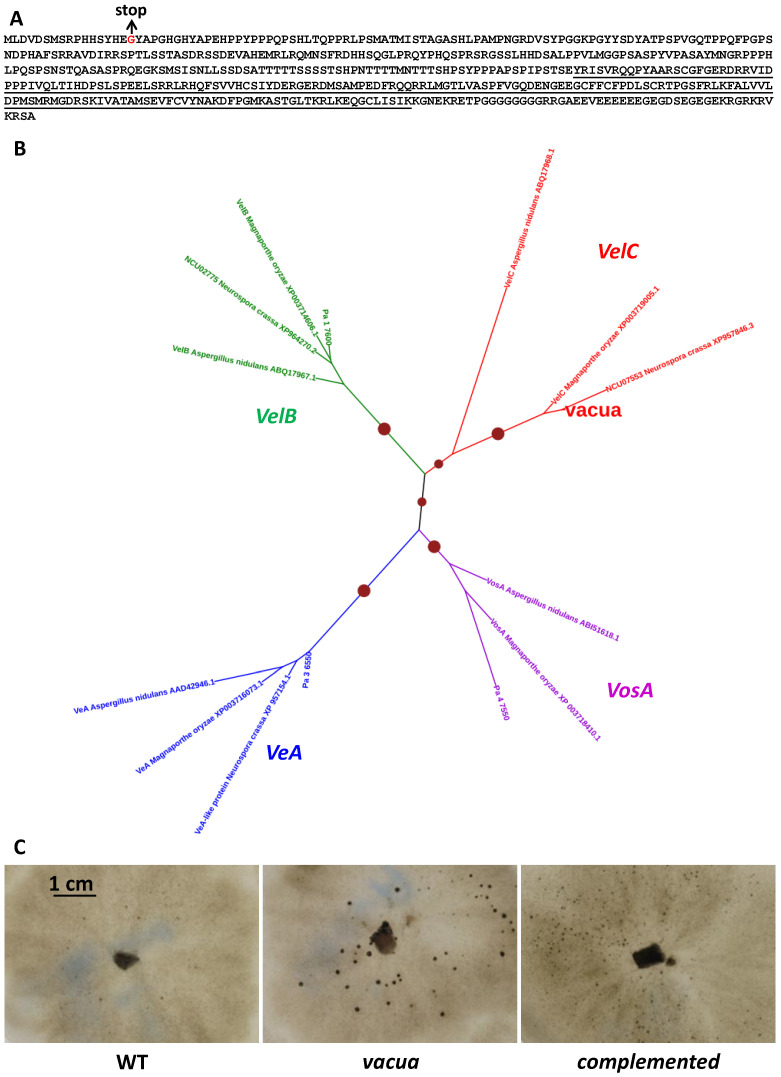
The previously described mutant vacua [12] presents a phenotype similar to pfd mutants, i.e., the production of normal-looking barren perithecia without fertilization. As for the mcm1 mutants, vacua did not produce barren fruiting bodies on G medium, in the dark and at 30 °C. However, the vacua mutation is not linked to the mat locus, indicating that it is not allelic with the MCM1 mutations. To identify the mutation responsible for the vacua phenotype, the genome of the mutant was sequenced. Beforehand, the vacua mutant was backcrossed ten times to the S strain, because it was obtained in the s strain [12] that contains numerous polymorphisms with respect to the S strain. Sequence analysis revealed a single mutation that could potentially account for the vacua phenotype: a G to T transversion changing codon 16 GGA coding for glycine into a TGA stop codon in the Pa_7_6330 CDS (Figure 8A). This suggested that the mutation had created a null allele. To ascertain that this mutation was the one responsible for the phenotype, the vacua mutant was co-transformed with the wild-type allele of Pa_7_6330 and a plasmid carrying a hygromycin B resistance gene. Among the 17 recovered hygromycin B-resistant transformants, one regained a wild-type phenotype, i.e., did not differentiate large perithecia in the absence of fertilization (Figure 8C). This transformant was crossed to the wild type. Analysis of the progeny indicated that restoration of a wild-type phenotype and hygromycin B resistance co-segregated, and that integration had occurred at a locus unlinked to vacua, showing that the mutation in Pa_7_6330 was indeed responsible for the vacua phenotype. Nevertheless, complementation was not complete since the complemented F0 and F1 strains differentiated small neckless protoperithecia that were slightly larger than those produced by the wild type (Figure 8C). Pa_7_6330 was thus renamed vacua. It encodes for a 55-kd protein that contain a velvet domain (Figure 8A). Phylogenetic analysis involving vacua and its homologues in P. anserina and related fungi (Figure 8B) revealed that vacua encoded a protein orthologous to the VelC genes of other fungi and that P. anserina contained a full complement of velvet genes (VeA = Pa_3_6550, VelB = Pa_1_7600, VelC = vacua and VosA = Pa_4_7550), as previously described [38].
Figure 8.
The vacua gene encodes a velvet protein. (A). protein sequence of the Pa_7_6330 CDS. The velvet domain is underlined and the position of the mutation in the vacua mutant is indicated in red. (B). Phylogenetic tree of vacua and its homologues. The tree was constructed with PhyML using the WAG model. Dot sizes are proportional to bootstrap values. Proteins used were P. anserina VeA = Pa_3_6550, VelB = Pa_1_7600, VelC = vacua and VosA = Pa_4_7550, Neurospora crassa Ve1 protein (= VeA- like protein; GenBank accession n° XP_957154.1), Ve2 (=VelB, CDS number NCU02775), and a protein similar to VelC (CDS number NCU07553), Magnaporthe grisea VeA (GenBank accession n° XP_003716073.1), VelB (GenBank accession n° XP_003714606.1), VelC (GenBank accession n° XP_003719005.1) and VosA (GenBank accession n° XP_003718410.1) and A. nidulans VeA (GenBank accession n° AAD42946.1), VelB (GenBank accession n° ABQ17967.1), VelC (GenBank accession n° ABQ17968.1) and VosA (GenBank accession n° ABI51618.1), (C). Unlike the wild type (WT), the vacua mutant differentiated normal-looking barren perithecia without fertilization (mature-like sterile fruiting bodies). Introduction of a wild-type copy of the vacua gene prevented formation of mature-like perithecia (complemented strain); however, the complemented strain differentiated slightly larger neckless protoperithecia, suggesting that complementation was partial.
The double mutant mcm1Δ vacua was obtained by crosses. As the vacua allele did not have a selection marker, the vacua locus from candidate hygromycin-resistant spores (mcm1Δ) were sequenced, after amplification by PCR.
The double mutant mcm1Δvacua spontaneously differentiated perithecia, although in lower amounts than the vacua mutant when alone, indicating that the mutation in vacua was epistatic over the null mutations in mcm1 (Figure 9). Conversely, it produced almost no spermatia like the single mcm1Δ mutant.
Figure 9.
Heterokaryotic crosses involving the vacua and mcm1Δ mutants. (A). The pictures were taken 7 days after fertilization, at which time the fully mature perithecia expelled ascospores. Perithecia are visible as small dots in the close-up view. (B). Microscopic observations of perithecia and typical rosette of asci observed in S and vacua fertilized perithecia and paraphyses observed in the double mutant after 7 days of growth.
3.9. Interaction with ste12
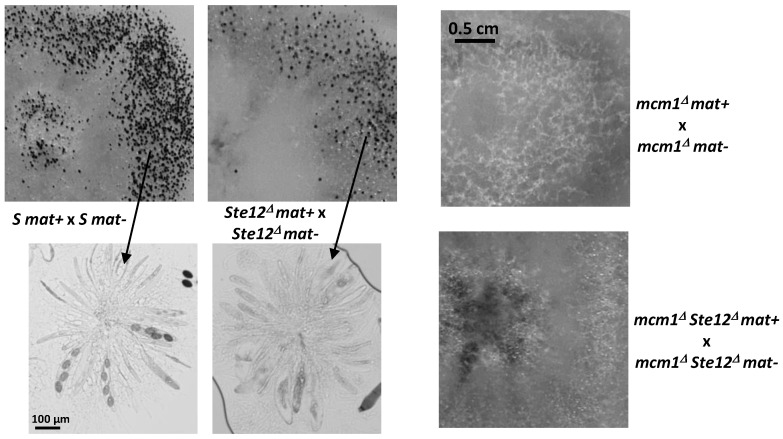
As in S. macrospora, MCM1 has been shown to interact with the homeodomain transcription factor (HD) Ste12 during ascospore formation [39], and therefore we analyzed the mcm1/ste12 interaction in P. anserina. To this end, a ste12∆ strain was constructed. This mutant presented a small delay in sexual reproduction (Figure 10 at day 7). It also featured a germination defect, as its germination rate was 58% instead of the 97% typical for the wild type.
Figure 10.
Heterokaryotic crosses involving the ste12Δ and mcm1Δ mutants. The pictures were taken 7 days after seeding, at which time fully mature perithecia expelled ascospores. Perithecia are visible as small dots in the close-up view. Below pointed by arrows microscopic observations of perithecia and typical rosette of asci observed in S perithecia and in Ste12∆ perithecia, showing a small delay in sexual development.
The double mutant was obtained in the progeny of mcm1Δ x ste12∆ crosses. It presented the same phenotype as the single mcm1Δ mutant alone, i.e., the double mutant was sterile, indicating that mutations in mcm1 were epistatic over mutations in ste12.
4. Discussion
Like in other fungi, perithecium differentiation in P. anserina is triggered by a proper set of environmental cues, e.g., light, temperature, adequate nutrients, etc., and proceeds mostly after fertilization, as seen in typical ascohymenial development. Indeed, the female gametangia stop their development as small protoperithecia and await fertilization. Upon fertilization, maturation of the fruiting bodies resumes and several tissues/specialized hyphae of maternal origin are then differentiated, including the neck, the paraphyses, the layers of the peridium, etc. This results in the mature perithecium that is several orders of magnitude larger than the protoperithecium. How development is coordinated to produce mature fruiting bodies properly ejecting ascospores is not fully understood. It was known from the recovery of the vacua mutant that maternal tissues can alone produce normal-looking, yet barren, perithecia, suggesting that the genetic maternal program(s) used to construct the fruiting bodies can be activated in the absence of fertilization. This advocates that ascohymenial and ascolocular developmental programs are not radically different and only differ in the timing of activation of these autonomous program(s) managing the development of the maternal tissue of the fruiting body. With the aim to understand how these program(s) are activated, we selected additional mutants with a phenotype similar to vacua. Identification of the genes affected in vacua and these newly isolated mutants should then help us to understand how development is initiated at the molecular level. To the best of our knowledge, only one other mutant from Magnaporthe grisea displays a similar phenotype, and the affected gene in this mutant is still unknown [40]. Note that such mutants can only be easily recovered in heterothallics or pseudohomothallics like P. anserina and M. grisea and not in homothallics such as S. macrospora and A. nidulans, in which many studies aimed at understanding the development of fruiting bodies are performed (see below).
Here, the identification of the genes affected in the newly-isolated pfd3, pfd9, and pfd23 mutants and of the P. anserina vacua mutant showed the involvement of the mcm1 and VelC genes in activating the development of perithecia. In these mutants, the ability to produce spontaneous perithecia is due solely to a bypass of fertilization, because they do not produce perithecia if the proper conditions are not met, i.e., the mutants still require availability of proper nutrients, an adequate temperature lower than 30 °C, and the presence of light to produce fruiting bodies.
The yeast MCM1 protein and its orthologues [14] belong to the MADS box protein family, with four other founding members, i.e., Arg80 (in yeast) [41], Agamous (in plants) [42], Deficiens (in plants) [43] and SRF (in humans) [44]; all contain a conserved DNA binding and dimerization domain named the MADS box. These proteins, described as cellular coordinators, are transcription factors present in most eukaryotic organisms. They control different developmental processes in plants, fungi, and animals. In animal and fungi, there are two distinct types of MADS box proteins, the SRF-like (type I) and MEF2-like (type II) classes [45]. MCM1 belongs to the SRF-like class. In S. cerevisiae, it was demonstrated to be involved in cell cycle control. This transcriptional factor is activated by the Bck2 protein and then induces expression of G1/S genes [46]. In this work, we showed that the MCM1 protein fused with GFP is localized in the nuclei of P. anserina. This is consistent with the prediction that the MCM1 protein is a transcription factor. In S. macrospora, as shown here in P. anserina, MCM1 controls different development processes and is essential for the formation of perithecia [47,48]. A similar role has been evidenced in A. nidulans [49], Fusarium graminearum [50], and Colletotrichun fructicola [51]. In the phytopathogen M. oryzae, the gene MoMCM1 was shown to be required for appressorial penetration and male fertility, but it does not seem to be involved in fruiting body formation [52]. The role of MCM1 has also been investigated by gene deletion in Beauveria bassiana [53] and Verticillium dahlia, [54], but its involvement in fruiting body formation has not been assayed.
Interestingly, phenotypes are different in the pfd point mutants and in the mcm1Δ deleted strain. The pfd9 mat- mutant, which has a mutation in the sequence encoding the MADS box domain, can form perithecia spontaneously, i.e., without fertilization. The same phenotype is observed in the pfd23 mat+ mutant, which has a shift in the open reading frame at the end of the coding sequence. On the other hand, the null pfd3 mat- mutant, which has a stop codon at the 8th codon, does not make spontaneous perithecia on its own. To observe such a phenotype, this mutant must be in heterokaryons with the wild-type strain. The same observations are made in mcm1Δ, where the mcm1 gene has been deleted. Of importance, the null mutants (pfd3 and mcm1Δ) are male and female sterile, without production of male or female gametes, whereas the point mutations in the pfd9 and pfd23 mutants do not appear to have strong effect on fertility. To account for this bizarre behavior, it can be hypothesized that the mutations present in the mcm1 gene in the pfd9 and pfd23 mutants result in partially functional MCM1 proteins. These would fail to repress properly the commitment of protoperithecia to continue their development, hence the spontaneous appearance of barren perithecia; however, they would be functional enough to allow gamete production and the subsequent steps of the maturation of the fruiting bodies. The MCM1 would thus have two functions: the first one as a repressor of the maternal program(s) enabling the making of the maternal tissues of the fruiting bodies while the maternal gametes wait for fertilizing male gametes, and the second one as an activator permitting fruiting body formation. What programs are repressed and activated is not yet clear. MCM1 is required in P. anserina for the production of spermatia and ascogonia. Failure to produce perithecia in the null mutants when no wild-type mcm1 nucleus is present could thus result solely from lack of gamete production. However, in S. macrospora, it has been shown that MCM1 is able to form homodimers and to interact with the mating-type protein SMTA-1, and then may be involved in the transcriptional regulation of mating-type-specific genes [47]; hence, MCM1 is likely required for “fertilization” in this homothallic fungus. This role may be conserved in P. anserina. It can be hypothesized that when wild-type nuclei are present, enough MCM1 protein would be supplied and diffuse into mutant hyphae to enable gamete production and/or fertilization. Once this critical step controlled by MCM1 is completed, perithecium maturation would continue because MCM1, whose gene is missing in such gametes, could not repress the development of maternal tissue.
As stated above, it has previously been shown that MCM1 interacts with the HD protein STE12, a transcription factor that activates genes of the mating pathway in yeast [39]. STE12 acts downstream of the MAP kinase FUS3/KSS1 cascade, where it plays an important role in mating and filamentation [55]. In A. nidulans, Neurospora crassa, and S. macrospora, STE12 also controls sexual development [39,56,57]. In S. macrospora, deletion of the ste12 genes induces a defect in ascospore formation, while in N. crassa and A. nidulans the mutants fail to produce protoperithecia/cleistothecia. In M. grisea, the STE12 mutants deleted for the Mst12 gene are both male and female fertile [58]; however, homozygous crosses of the STE12 mutants were not performed, preventing us from knowing whether STE12 is required for perithecium development, especially in the knowledge that, as in S. macrospora, MoMCM1 interacts with both Mst12 and Mata-1 [52]. In P. anserina, we show that the STE12 ortholog has a small role during sexual reproduction, since homozygous crosses of the mutant were fertile and only slightly delayed when compared with the wild type, the major defect being an impairment in ascospore germination. As would be expected from their phenotypes, the double mutant mcm1Δ ste12∆presented the same phenotype as the single mcm1Δ mutant alone, suggesting that there is no interaction between Ste12 and MCM1 during the developmental process in P. anserina.
The role of the VelC gene has been investigated in A. nidulans, M. grisea, and Valsa mali [59], but its role in fertility was only assayed in A. nidulans, where it was shown that its deletion results in diminished production of cleistothecia, while its overexpression has the opposite effect [60]). In N. crassa, the VelC orthologue “VE-3” is described as being non-functional [61] and its deletion appears to have no effect (see the unpublished data mentioned in [38]). However, upon examination of the genome sequences of strain OR74, it appears that VE-3 is badly annotated (see Supporting Information Figure S3), because an additional base is present within the coding sequence, creating a frameshift. This triggers the annotation to predict a false intron and the lack of part of the velvet domain in the predicted VE-3 protein. The supplementary base is not present in the genome sequence of strain ORS-SL6a; hence, in this N. crassa strain, VE-3 appears fully functional and its role remains to be assessed. Here, we show that it is involved in P. anserina in controlling fruiting body development, as in A. nidulans. Based on the phenotype of the mcm1Δvacua double mutant, it is likely that MCM1 and the vacua proteins operate either in independent pathways to bypass fertilization or more likely that vacua acts downstream of MCM1 in the same signaling pathway. Unlike the other members of the velvet family [61], there is at present no evidence to suggest that vacua/Velc is involved in light regulation as the vacua mutant did not produce mature fruiting bodies in the dark.
Phenotypic characterization continued by testing different types of stress on the mutants in this study. In particular, a previous study on the yeast S. cerevisiae showed that MCM1 is required for transcription of a subset of genes involved in maintenance of the cell wall and then is involved in resistance to calcofluor, a compound that binds to chitin in fungal cells [36]. The deleted strains, as well as the pfd3 and pfd23 mutants, appeared to be more resistant to calcofluor than the wild-type strain, which is the reverse of the phenotypes observed in yeast. The pfd9 mutant showed a phenotype close to that of the wild-type strain, which would confirm that the MCM1 protein is partially functional in this strain. Tests with Congo red, a glucan synthase inhibitor, showed similar results. Further experiments must be undertaken to understand how MCM1 is involved in the regulation of fungal wall formation. Moreover, contrary to what is observed in S. cerevisiae [62], the MCM1 protein does not seem to be involved in osmotic stress resistance in P. anserina. Finally, none of the mutants tested showed any particular phenotype with respect to oxidative stress or growth at low temperature (18 °C). At 35 °C, it has been described that the S mat- strain is thermoresistant [63]. The pfd3 mat- and mcm1Δ mat- mutants tested in this study both became thermosensitive at 35 °C. Thus, it appears that the MCM1 transcription factor directly or indirectly regulates the expression of the RMP gene, which is responsible for the 35 °C thermoresistance of the S mat- strain. It is interesting to note that the RMP gene is, like the mcm1 gene, linked to the mat locus.
Antifungal testing on the different strains showed an increase in resistance to azole antifungals in link with deletion or mutations in the mcm1 gene. These results are contradictory to the results obtained in C. albicans [64], in which deletion of mcm1 caused hypersensitivity of strains to azoles. In C. albicans, it has been shown that azoles can be expelled normally from the cell through activation of MDR1 efflux pumps via the MCM1 transcription factor [64]. Then, it has been hypothesized that, when mcm1 is deleted, pumps are no longer activated, and the antifungals are no longer properly expelled, which results in hypersensitivity to azole antifungals. In this study, P. anserina mutants appear to be more resistant to azole antifungals and to a lesser extent to polyene antifungals (amphotericin B). It is conceivable that, in this case, the transcription factor represses efflux pump expression in the S strain and that inactivation of mcm1 causes deregulation of efflux pump expression or constitutive expression, resulting in higher resistance to antifungal drugs.
5. Conclusions
In conclusion, classical genetic analysis has permitted us to identify the MCM1 and VelC proteins as crucial for repressing maternal tissue development in P. anserina. According to the results provided by this study, the mcm1 gene in P. anserina probably encodes a transcription factor, as suggested by its nuclear localization. In addition to repressing maternal tissue development, it also controls the production of both male and female gametes. Analysis of the interactions between the mcm1 and vacua mutants tentatively suggests that mcm1 and vacua act in the same pathway, with vacua acting downstream of mcm1.
Acknowledgments
We warmly thank Sylvie Cangemi and Aurélien Renault for their expert technical assistance. Insaf Essadik was supported by a PhD scholarship from the Doctoral School MTCI (ED 563).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof10010079/s1, Supporting Information Figure S1. Southern blot analysis for final validation. Supporting Information Figure S2. Methodological approach for perithecia enumeration and area estimation made by Aphelion Imaging Software. Supporting Information Figure S3. Sequences of the VelC homologues of N. crassa in strains ORS-SL6a and OR74A. The additional base in OR74A is boxed in white. Bottom, domain analysis using CD-search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 4 December 2023) of the VelC homologue of ORS-SL6a showing that in this strain the velvet domain is complete. Supporting Information Table S1. Primers used for mcm1 (Pa_1_19280), ste12 (Pa-7-1730) and vacua (Pa-7-6330) gene deletion, complementation, and detection. Supporting Information Table S2: Summarize of global measurements, based on perithecia surfaces (mm2); Count: total number of extracted objects for each replicate; area_mean and area_med are the mean value and median value of perithecia for each replicate; areas; std is the standard deviation of area datasets.
Author Contributions
Conceptualization, P.S., F.C.-L. and G.R.-R.; methodology, I.E., C.B. (Charlie Boucher), C.B. (Cécilia Bobée), É.C., V.G., H.L, P.S. and G.R.-R.; validation, I.E., C.B. (Charlie Boucher), C.B. (Cécilia Bobée), É.C., V.G., H.L. and P.S.; formal analysis, C.B. (Cécilia Bobée), É.C., F.C.-L. and G.R.-R.; investigation, I.E., C.B. (Charlie Boucherand), C.B. (Cécilia Bobée), É.C., V.G, P.S. and G.R.-R.; resources, I.E., C.B. (Charlie Boucherand), C.B. (Cécilia Bobée), É.C., V.G.; writing—original draft preparation, F.C.-L. and G.R.-R.; writing—review and editing, P.S., F.C.-L. and G.R.-R.; visualization, P.S., C.B. (Cécilia Bobée), É.C., F.C.-L. and G.R.-R.; supervision, F.C.-L. and G.R.-R.; project administration, F.C.-L. and G.R.-R.; funding acquisition, P.S., F.C.-L. and G.R.-R. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Data are contained within the article and supplementary materials.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This work was supported by an intramural funding from Université Paris 7 (now included in the Université Paris Cité) and a grant P3AMB from Region Ile de France.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Peraza-Reyes L., Berteaux-Lecellier V. Peroxisomes and sexual development in fungi. Front. Physiol. 2013;4:244. doi: 10.3389/fphys.2013.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silar P. Simple Genetic Tools to Study Fruiting Body Development in Fungi. Open Mycol. J. 2014;8:148–155. doi: 10.2174/1874437001408010148. [DOI] [Google Scholar]
- 3.Dyer P.S., O'Gorman C.M. Sexual development and cryptic sexuality in fungi: Insights from Aspergillus species. FEMS Microbiol. Rev. 2012;36:165–192. doi: 10.1111/j.1574-6976.2011.00308.x. [DOI] [PubMed] [Google Scholar]
- 4.Chinnici J.L., Fu C., Caccamise L.M., Arnold J.W., Free S.J. Neurospora crassa Female Development Requires the PACC and Other Signal Transduction Pathways, Transcription Factors, Chromatin Remodeling, Cell-To-Cell Fusion, and Autophagy. PLoS ONE. 2014;9:e110603. doi: 10.1371/journal.pone.0110603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teichert I., Nowrousian M., Pöggeler S., Kück U. The Filamentous Fungus Sordaria Macrospora as a Genetic Model to Study Fruiting Body Development. Adv. Genet. 2014;87:199–244. doi: 10.1016/B978-0-12-800149-3.00004-4. [DOI] [PubMed] [Google Scholar]
- 6.Pöggeler S., Nowrousian M., Teichert I., Beier A., Kück U. Fruiting-Body Development in Ascomycetes. In: Anke T., Schüffler A., editors. Physiology and Genetics: Selected Basic and Applied Aspects. Springer International Publishing; Cham, Switzerland: 2018. pp. 1–56. The Mycota. [Google Scholar]
- 7.Debuchy R., Berteaux-Lecellier V., Silar P. Mating Systems and Sexual Morphogenesis in Ascomycetes. Cell. Mol. Biol. Filamentous Fungi. 2010;14:501–535. doi: 10.1128/9781555816636.ch33. [DOI] [Google Scholar]
- 8.Nowrousian M. Genomics and transcriptomics to study fruiting body development: An update. Fungal Biol. Rev. 2018;32:231–235. doi: 10.1016/j.fbr.2018.02.004. [DOI] [Google Scholar]
- 9.Lütkenhaus R., Traeger S., Breuer J., Carreté L., Kuo A., Lipzen A., Pangilinan J., Dilworth D., Sandor L., Pöggeler S., et al. Comparative Genomics and Transcriptomics To Analyze Fruiting Body Development in Filamentous Ascomycetes. Genetics. 2019;213:1545–1563. doi: 10.1534/genetics.119.302749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lalucque H., Malagnac F., Green K., Gautier V., Grognet P., Tong L.C.H., Scott B., Silar P. IDC2 and IDC3, two genes involved in cell non-autonomous signaling of fruiting body development in the model fungus Podospora anserina. Dev. Biol. 2017;421:126–138. doi: 10.1016/j.ydbio.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 11.Silar P. Podospora anserina: From Laboratory to Biotechnology. In: Horwitz B.A., Mukherjee P.K., Mukherjee M., Kubicek C.P., editors. Genomics of Soil- and Plant-Associated Fungi. Springer; Berli/Heidelberg, Germany: 2013. pp. 283–309. (Soil Biology). [Google Scholar]
- 12.Esser K., Graw D. Homokaryotic Fruiting in the Bipolar-Incompatible Ascomycete Podospora anserina. Mycologia. 1980;72:534–541. doi: 10.1080/00275514.1980.12021215. [DOI] [Google Scholar]
- 13.Passmore S., Maine G.T., Elble R., Christ C., Tye B.-K. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MATα cells. J. Mol. Biol. 1988;204:593–606. doi: 10.1016/0022-2836(88)90358-0. [DOI] [PubMed] [Google Scholar]
- 14.Passmore S., Elble R., Tye B.K. A protein involved in minichromosome maintenance in yeast binds a transcriptional enhancer conserved in eukaryotes. Minerva Anestesiol. 1989;3:921–935. doi: 10.1101/gad.3.7.921. [DOI] [PubMed] [Google Scholar]
- 15.Espagne E., Lespinet O., Malagnac F., Da Silva C., Jaillon O., Porcel B.M., Couloux A., Aury J.-M., Ségurens B., Poulain J., et al. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 2008;9:R77. doi: 10.1186/gb-2008-9-5-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silar P. Podospora anserina. 2020. [(accessed on 4 December 2023)]. Available online: https://hal.archives-ouvertes.fr/hal-02475488.
- 17.El-Khoury R., Sellem C.H., Coppin E., Boivin A., Maas M.F.P.M., Debuchy R., Sainsard-Chanet A. Gene deletion and allelic replacement in the filamentous fungus Podospora anserina. Curr. Genet. 2008;53:249–258. doi: 10.1007/s00294-008-0180-3. [DOI] [PubMed] [Google Scholar]
- 18.Coppin E., Silar P. Identification of PaPKS1, a polyketide synthase involved in melanin formation and its use as a genetic tool in Podospora anserina. Mycol. Res. 2007;111:901–908. doi: 10.1016/j.mycres.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Grognet P., Bidard F., Kuchly C., Tong L.C.H., Coppin E., Benkhali J.A., Couloux A., Wincker P., Debuchy R., Silar P. Maintaining Two Mating Types: Structure of the Mating Type Locus and Its Role in Heterokaryosis in Podospora anserina. Genetics. 2014;197:421–432. doi: 10.1534/genetics.113.159988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H. Improving SNP discovery by base alignment quality. Bioinformatics. 2011;27:1157–1158. doi: 10.1093/bioinformatics/btr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koboldt D.C., Larson D.E., Wilson R.K. Using VarScan 2 for Germline Variant Calling and Somatic Mutation Detection. Curr. Protoc. Bioinform. 2013;44:15.4.1–15.4.17. doi: 10.1002/0471250953.bi1504s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grigoriev I.V., Nikitin R., Haridas S., Kuo A., Ohm R., Otillar R., Riley R., Salamov A., Zhao X., Korzeniewski F., et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2013;42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katoh K., Kuma K.I., Toh H., Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 28.Letunic I., Bork P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 29.Lalucque H., Malagnac F., Brun S., Kicka S., Silar P. A Non-Mendelian MAPK-Generated Hereditary Unit Controlled by a Second MAPK Pathway in Podospora anserina. Genetics. 2012;191:419–433. doi: 10.1534/genetics.112.139469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bidard F., Benkhali J.A., Coppin E., Imbeaud S., Grognet P., Delacroix H., Debuchy R. Genome-Wide Gene Expression Profiling of Fertilization Competent Mycelium in Opposite Mating Types in the Heterothallic Fungus Podospora anserina. PLoS ONE. 2011;6:e21476. doi: 10.1371/journal.pone.0021476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coppin E., Berteaux-Lecellier V., Bidard F., Brun S., Ruprich-Robert G., Espagne E., Aït-Benkhali J., Goarin A., Nesseir A., Planamente S., et al. Systematic Deletion of Homeobox Genes in Podospora anserina Uncovers Their Roles in Shaping the Fruiting Body. PLoS ONE. 2012;7:e37488. doi: 10.1371/journal.pone.0037488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silar P. Two new easy to use vectors for transformations. Fungal Genet. Rep. 1995;42:73. doi: 10.4148/1941-4765.1353. [DOI] [Google Scholar]
- 33.Tong L.C.H., Silar P., Lalucque H. Genetic control of anastomosis in Podospora anserina. Fungal Genet. Biol. 2014;70:94–103. doi: 10.1016/j.fgb.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Brun S., Malagnac F., Bidard F., Lalucque H., Silar P. Functions and regulation of the Nox family in the filamentous fungus Podospora anserina: A new role in cellulose degradation. Mol. Microbiol. 2009;74:480–496. doi: 10.1111/j.1365-2958.2009.06878.x. [DOI] [PubMed] [Google Scholar]
- 35.Jamet-Vierny C., Debuchy R., Prigent M., Silar P. IDC1, a Pezizomycotina-specific gene that belongs to the PaMpk1 MAP kinase transduction cascade of the filamentous fungus Podospora anserina. Fungal Genet. Biol. 2007;44:1219–1230. doi: 10.1016/j.fgb.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Abraham D.S., Vershon A.K. N-Terminal Arm of Mcm1 Is Required for Transcription of a Subset of Genes Involved in Maintenance of the Cell Wall. Eukaryot. Cell. 2005;4:1808–1819. doi: 10.1128/EC.4.11.1808-1819.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Messenguy F., Dubois E. Role of MADS Box Proteins and Their Cofactors in Combinatorial Control of Gene Ex-pression and Cell Development. Gene. 2003;316:1–21. doi: 10.1016/S0378-1119(03)00747-9. [DOI] [PubMed] [Google Scholar]
- 38.Ojeda-López M., Chen W., Eagle C.E., Gutiérrez G., Jia W.L., Swilaiman S.S., Huang Z., Park H.-S., Yu J.-H., Cánovas D., et al. Evolution of asexual and sexual reproduction in the Aspergilli. Stud. Mycol. 2018;91:37–59. doi: 10.1016/j.simyco.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nolting N., Pöggeler S. A STE12 homologue of the homothallic ascomycete Sordaria macrospora interacts with the MADS box protein MCM1 and is required for ascosporogenesis. Mol. Microbiol. 2006;62:853–868. doi: 10.1111/j.1365-2958.2006.05415.x. [DOI] [PubMed] [Google Scholar]
- 40.Tharreau D., Notteghem J.-L., Lebrun M.-H. Mutations Affecting Perithecium Development and Sporulation in Magnaporthe grisea. Fungal Genet. Biol. 1997;21:206–213. doi: 10.1006/fgbi.1996.0951. [DOI] [Google Scholar]
- 41.Dubois E., Bercy J., Messenguy F. Characterization of two genes, ARGRI and ARGRIII required for specific regulation of arginine metabolism in yeast. Mol. Genet. Genom. 1987;207:142–148. doi: 10.1007/BF00331501. [DOI] [PubMed] [Google Scholar]
- 42.Yanofsky M.F., Ma H., Bowman J.L., Drews G.N., Feldmann K.A., Meyerowitz E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- 43.Sommer H., Beltrán J.P., Huijser P., Pape H., Lönnig W.E., Saedler H., Schwarz-Sommer Z. Deficiens, a Homeotic Gene Involved in the Control of Flower Morphogenesis in Antirrhinum majus: The Protein Shows Homology to Transcrip-tion Factors. EMBO J. 1990;9:605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norman C., Runswick M., Pollock R., Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55:989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- 45.Shore P., Sharrocks A.D. The MADS-Box Family of Transcription Factors. Eur. J. Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 46.Bastajian N., Friesen H., Andrews B.J. Bck2 Acts through the MADS Box Protein Mcm1 to Activate Cell-Cycle-Regulated Genes in Budding Yeast. PLoS Genet. 2013;9:e1003507. doi: 10.1371/journal.pgen.1003507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nolting N., Pöggeler S. A MADS Box Protein Interacts with a Mating-Type Protein and Is Required for Fruiting Body Development in the Homothallic Ascomycete Sordaria macrospora. Eukaryot. Cell. 2006;5:1043–1056. doi: 10.1128/EC.00086-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elleuche S., Bernhards Y., Schäfers C., Varghese J.M., Nolting N., Pöggeler S. The small serine-threonine protein SIP2 interacts with STE12 and is involved in ascospore germination in Sordaria macrospora. Eur. J. Cell Biol. 2010;89:873–887. doi: 10.1016/j.ejcb.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 49.Li N., Kunitake E., Endo Y., Aoyama M., Kanamaru K., Kimura M., Kato M., Kobayashi T. Involvement of an SRF-MADS protein McmA in regulation of extracellular enzyme production and asexual/sexual development in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2016;80:1820–1828. doi: 10.1080/09168451.2016.1146074. [DOI] [PubMed] [Google Scholar]
- 50.Yang C., Liu H., Li G., Liu M., Yun Y., Wang C., Ma Z., Xu J. The MADS-box transcription factor FgMcm1 regulates cell identity and fungal development in Fusarium graminearum. Environ. Microbiol. 2015;17:2762–2776. doi: 10.1111/1462-2920.12747. [DOI] [PubMed] [Google Scholar]
- 51.Liu W., Han L., Chen J., Liang X., Wang B., Gleason M.L., Zhang R., Sun G. The CfMcm1 Regulates Pathogenicity, Conidium Germination, and Sexual Development in Colletotrichum fructicola. Phytopathology®. 2022;112:2159–2173. doi: 10.1094/PHYTO-03-22-0090-R. [DOI] [PubMed] [Google Scholar]
- 52.Zhou X., Liu W., Wang C., Xu Q., Wang Y., Ding S., Xu J. A MADS-box transcription factor MoMcm1 is required for male fertility, microconidium production and virulence in Magnaporthe oryzae. Mol. Microbiol. 2011;80:33–53. doi: 10.1111/j.1365-2958.2011.07556.x. [DOI] [PubMed] [Google Scholar]
- 53.Zhao X., Yang X., Lu Z., Wang H., He Z., Zhou G., Luo Z., Zhang Y. MADS-box transcription factor Mcm1 controls cell cycle, fungal development, cell integrity and virulence in the filamentous insect pathogenic fungus Beauveria bassiana. Environ. Microbiol. 2018;21:3392–3416. doi: 10.1111/1462-2920.14629. [DOI] [PubMed] [Google Scholar]
- 54.Xiong D., Wang Y., Tian L., Tian C. MADS-Box Transcription Factor VdMcm1 Regulates Conidiation, Microsclerotia Formation, Pathogenicity, and Secondary Metabolism of Verticillium dahliae. Front. Microbiol. 2016;7:1192. doi: 10.3389/fmicb.2016.01192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chou S., Lane S., Liu H. Regulation of Mating and Filamentation Genes by Two Distinct Ste12 Complexes in Saccharomyces cerevisiae. Mol. Cells Biol. 2006;26:4794–4805. doi: 10.1128/MCB.02053-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vallim M.A., Miller K.Y., Miller B.L. Aspergillus stea (Sterile12-like) is a homeodomain-C2/H2-Zn+2 finger transcription factor required for sexual reproduction. Mol. Microbiol. 2000;36:290–301. doi: 10.1046/j.1365-2958.2000.01874.x. [DOI] [PubMed] [Google Scholar]
- 57.Li D., Bobrowicz P., Wilkinson H.H., Ebbole D.J. A Mitogen-Activated Protein Kinase Pathway Essential for Mating and Contributing to Vegetative Growth in Neurospora crassa. Genetics. 2005;170:1091–1104. doi: 10.1534/genetics.104.036772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park G., Xue C., Zheng L., Lam S., Xu J.-R. MST12 Regulates Infectious Growth But Not Appressorium Formation in the Rice Blast Fungus Magnaporthe grisea. Mol. Plant-Microbe Interact. 2002;15:183–192. doi: 10.1094/MPMI.2002.15.3.183. [DOI] [PubMed] [Google Scholar]
- 59.Kim H.-J., Han J.-H., Kim K.S., Lee Y.-H. Comparative functional analysis of the velvet gene family reveals unique roles in fungal development and pathogenicity in Magnaporthe oryzae. Fungal Genet. Biol. 2014;66:33–43. doi: 10.1016/j.fgb.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 60.Park H.-S., Nam T.-Y., Han K.-H., Kim S.C., Yu J.-H. VelC Positively Controls Sexual Development in Aspergillus nidulans. PLoS ONE. 2014;9:e89883. doi: 10.1371/journal.pone.0089883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bayram S., Dettmann A., Karahoda B., Moloney N.M., Ormsby T., McGowan J., Cea-Sánchez S., Miralles-Durán A., Brancini G.T.P., Luque E.M., et al. Control of Development, Secondary Metabolism and Light-Dependent Carotenoid Biosynthesis by the Velvet Complex of Neurospora crassa. Genetics. 2019;212:691–710. doi: 10.1534/genetics.119.302277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuo M.-H., Nadeau E.T., Grayhack E.J. Multiple Phosphorylated Forms of the Saccharomyces cerevisiae Mcm1 Protein Include an Isoform Induced in Response to High Salt Concentrations. Mol. Cells Biol. 1997;17:819–832. doi: 10.1128/MCB.17.2.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Contamine V., Zickler D., Picard M. The Podospora rmp1 Gene Implicated in Nucleus-Mitochondria Cross-Talk Encodes an Essential Protein Whose Subcellular Location Is Developmentally Regulated. Genetics. 2004;166:135–150. doi: 10.1534/genetics.166.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mogavero S., Tavanti A., Senesi S., Rogers P.D., Morschhäuser J. Differential Requirement of the Transcription Factor Mcm1 for Activation of the Candida albicans Multidrug Efflux Pump MDR1 by Its Regulators Mrr1 and Cap1. Antimicrob. Agents Chemother. 2011;55:2061–2066. doi: 10.1128/AAC.01467-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article and supplementary materials.