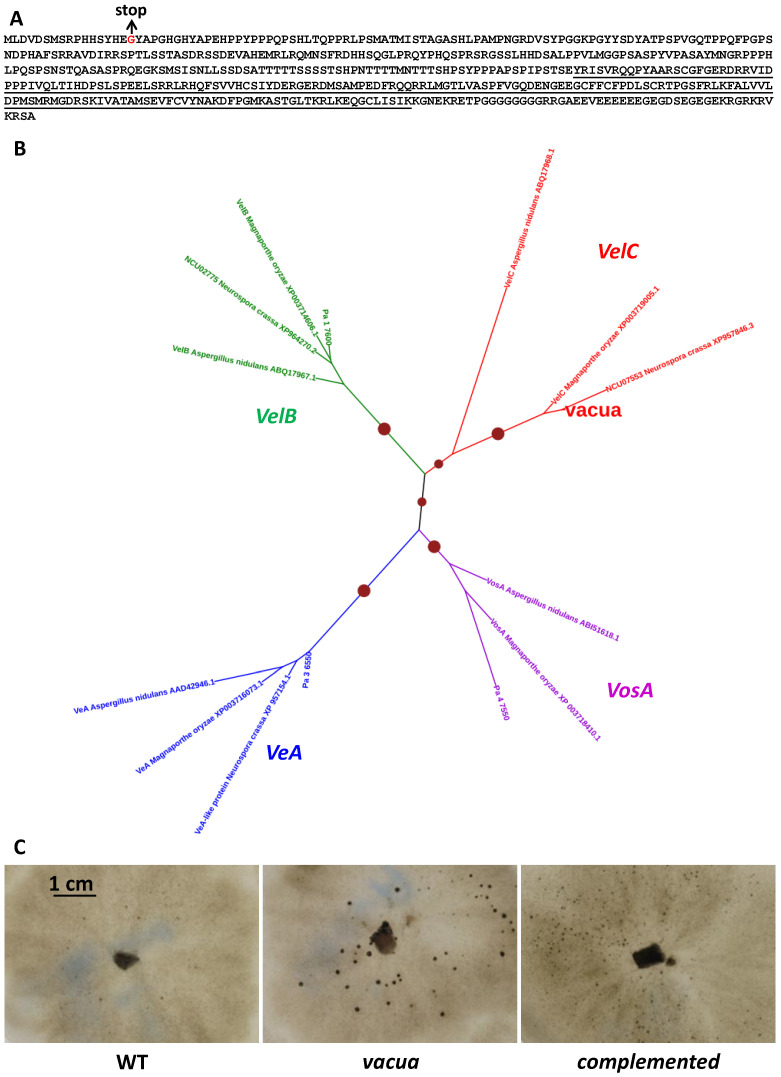
Figure 8.
The vacua gene encodes a velvet protein. (A). protein sequence of the Pa_7_6330 CDS. The velvet domain is underlined and the position of the mutation in the vacua mutant is indicated in red. (B). Phylogenetic tree of vacua and its homologues. The tree was constructed with PhyML using the WAG model. Dot sizes are proportional to bootstrap values. Proteins used were P. anserina VeA = Pa_3_6550, VelB = Pa_1_7600, VelC = vacua and VosA = Pa_4_7550, Neurospora crassa Ve1 protein (= VeA- like protein; GenBank accession n° XP_957154.1), Ve2 (=VelB, CDS number NCU02775), and a protein similar to VelC (CDS number NCU07553), Magnaporthe grisea VeA (GenBank accession n° XP_003716073.1), VelB (GenBank accession n° XP_003714606.1), VelC (GenBank accession n° XP_003719005.1) and VosA (GenBank accession n° XP_003718410.1) and A. nidulans VeA (GenBank accession n° AAD42946.1), VelB (GenBank accession n° ABQ17967.1), VelC (GenBank accession n° ABQ17968.1) and VosA (GenBank accession n° ABI51618.1), (C). Unlike the wild type (WT), the vacua mutant differentiated normal-looking barren perithecia without fertilization (mature-like sterile fruiting bodies). Introduction of a wild-type copy of the vacua gene prevented formation of mature-like perithecia (complemented strain); however, the complemented strain differentiated slightly larger neckless protoperithecia, suggesting that complementation was partial.