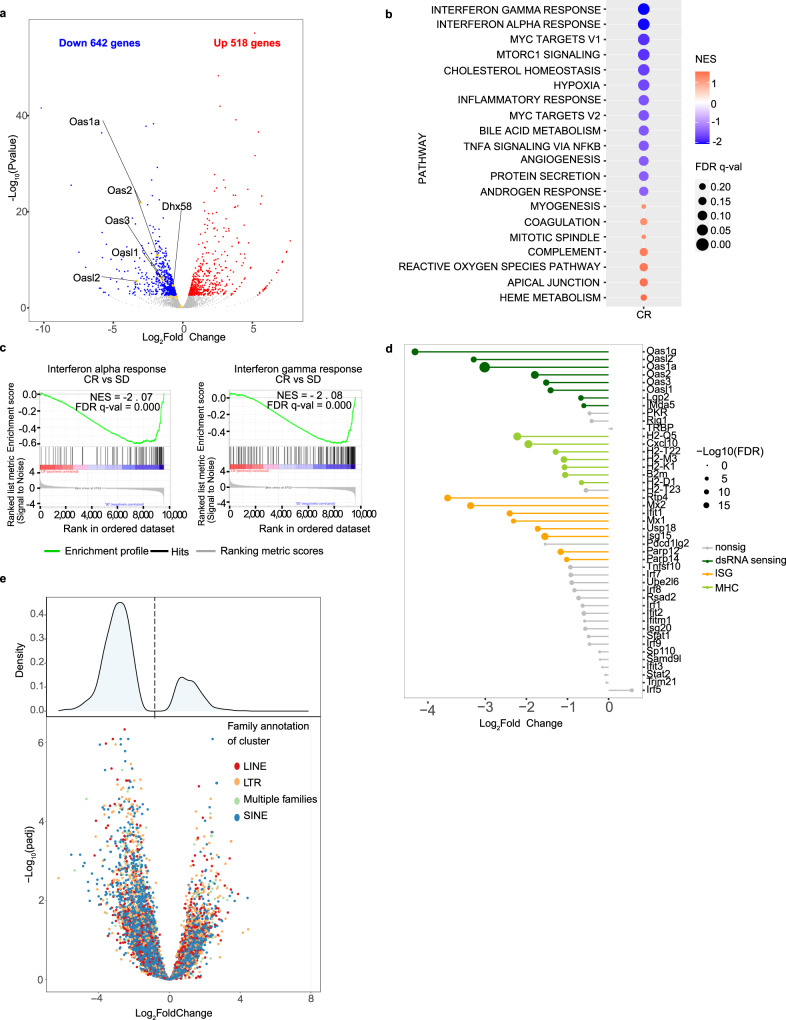
Fig. 2. Caloric restriction decreases the IFN/dsRNA signaling in the APL blasts.
a Volcano plot showing differentially expressed genes in CR and SD APL cells (RNAseq analyses from a pool of 2 mice per condition). Genes in red or blue are significantly up or down-regulated (FDR < 0.05). dsRNA sensing genes of interest are labeled. b hallmark GSEA terms enriched/depleted in CR and SD. The color of the dots indicates the normalized enrichment score (NES). The size indicates the FDR q-value. Note that only dots corresponding to pathways with an FDR < 0.25 are reported. c GSEA plots showing enrichment/depletion in CR vs SD of the interferon alpha and interferon gamma signatures, as indicated. d Differential expression of dsRNA sensing genes, MHC genes and other ISGs between CR and SD. Genes in gray(“nonsig”) are not significantly differentially expressed, with an FDR > 0.01. e Differential expression of ERV families from stranded RNAseq in CR vs. SD. Upper panel: density plot of log2 fold change; bottom panel: volcano plot of log2 fold change vs -log10 FDR. Colored dots are significantly deregulated genes at FDR < 0.05. Source data are provided as a Source Data file.