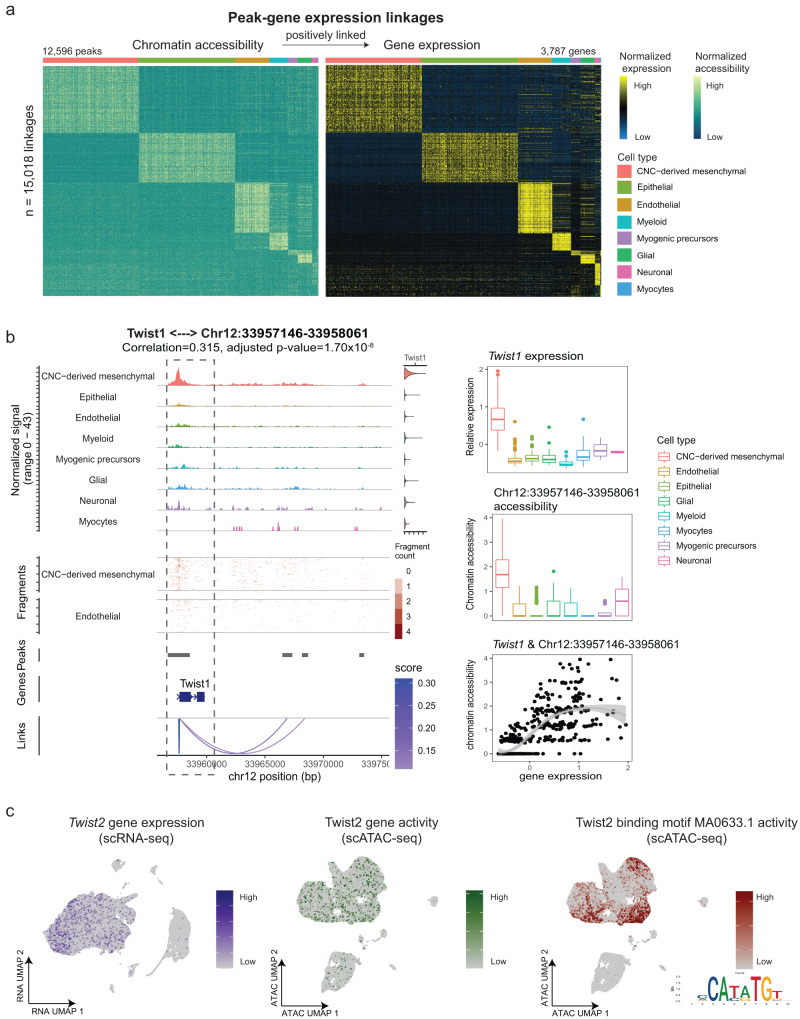
Fig. 2. Integration of chromatin accessibility with gene expression reveals cell-type-specific multiomic marker genes.
a Heatmap shows normalized chromatin accessibility (left) and gene expression (right) of 15,018 significantly linked peak-gene pairs. Each row represents a positively linked pair of regulatory elements and genes. Bar on the top represents major cell types. b Left: Genome Browser visualization of aggregated chromatic accessibility at the chr12: 33.96–33.97 (Mb) locus for each major cell type, coupled with Twist1 gene expression. Arcs at the bottom denote positively linked peak-Twist1 pairs. The linkage between Twist1 and chr12: 33,957,146–33,958,061 is highlighted with gray dotted box. The Pearson correlation test is employed for each pair of peak-gene, and multiple test adjustments are applied. Right: Boxplots show the expression level of Twist1 (top) and accessibility of the linked peak chr12: 33,957,146–33,958,061 (middle) across cell types. Minima: Lower limit of the whisker. Maxima: Upper limit of the whisker. Centre: Median line inside the box. The upper and lower box bounds represent the 25% and 75% percentile of data. The scatter plot shows the significant correlation between Twist1 expression and chr12: 33,957,146–33,958,061 accessibility (bottom). Each dot represents the aggregated gene expression and accessibility of ten cells from n = 28,529 biologically independent CNC-derived mesenchymal cells. The shaded areas around the fitted smooth line represent the error bands, indicating the 95% confidence interval. c UMAP visualizations illustrate the multimodal profiling of Twist2, including gene expression (left), gene activity (middle), and TWIST2 binding motif MA0633.1 activity (right). The position weight matrix of the MA0633.1 motif is embedded in the bottom right corner.