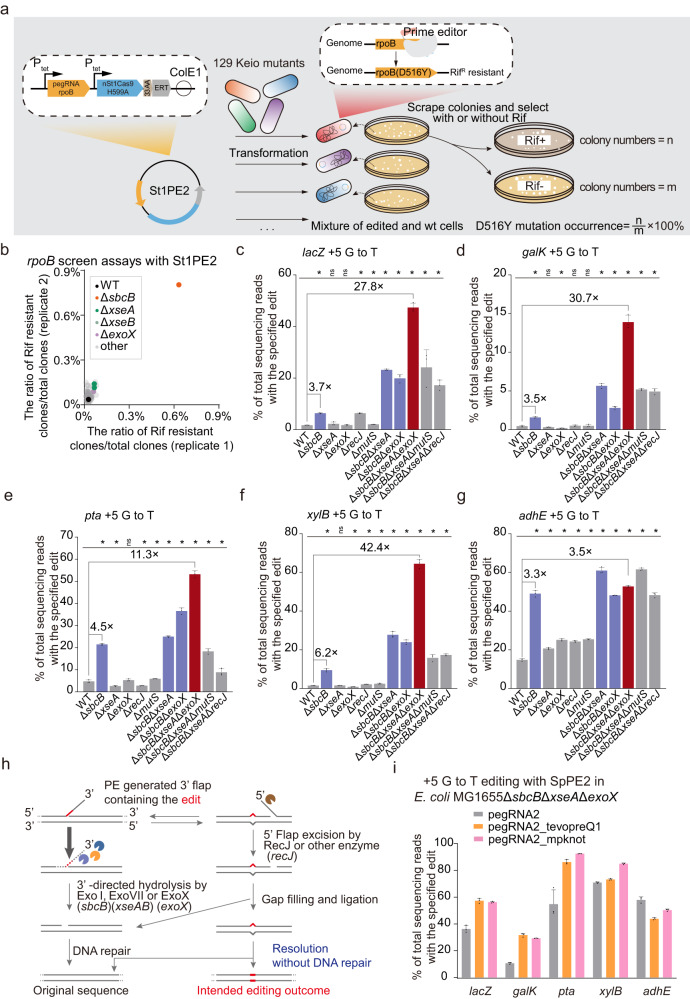
Fig. 2. Identification of key genetic determinates that impede prime editing in E. coli.
a Schematic of the rpoB-based genetic screening approach. 129 Keio mutants were individually transformed with St1PE2 to enable the conversion of D516Y mutation in rpoB. The ratio of (Rif resistant cononies)/(total colonies) was calculated by the number of colonies on Rif+ plate divided by the number of colonies on Rif- plate. b Identification of the key genes that inhibit prime editing using the rpoB-based screening assay. All values from n = 2 independent replicates are shown. c–g Comparison of the prime editing efficiency in E. coli MG1655 at different targeting loci lacZ (c), galK (d), pta (e), xylB (f), and adhE (g). ΔsbcB, ΔsbcBΔxseA, and ΔsbcBΔexoX mutants are colored blue, ΔsbcBΔxseAΔexoX mutants are colored red. The black stars represent the statistical differences between WT and mutants, respectively. Two-tailed student’s t test was performed. *p < 0.05. Data represent mean ± s.d. of n = 3 independent replicates. h The 3′-directed hydrolysis model that inhibits prime editing in E. coli. The prime editing intermediate was digested by exonucleases. i Impact of 3′ RNA structural motif on prime editing efficiency. Data represent mean ± s.d. of n = 3 independent replicates.