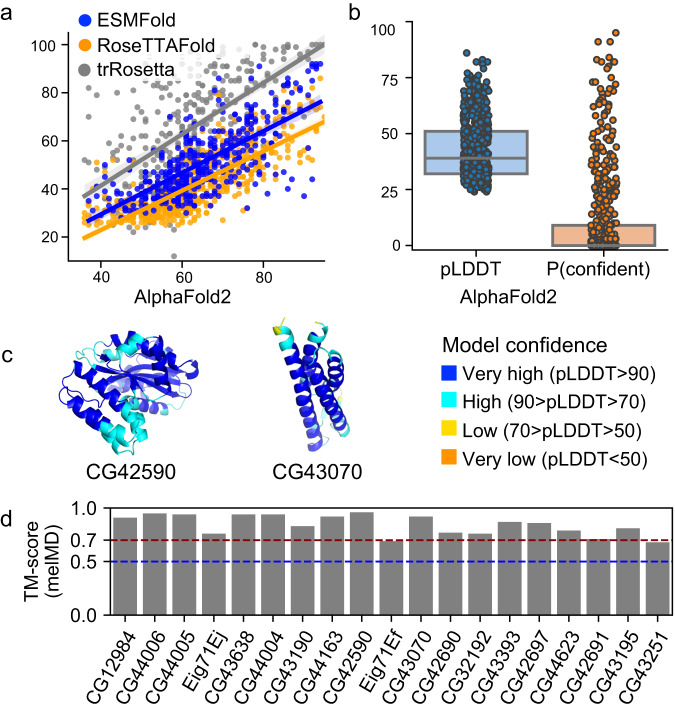
Fig. 3. AlphaFold2 structure predictions of de novo gene candidates.
a AlphaFold2 predictions are strongly correlated with RoseTTAFold and trRosetta predictions. The confidence interval is estimated by 1000 bootstrap resamples and is shown in shadow. b Most de novo gene candidates might not have well-folded protein structures according to pLDDT, per-residue confidence score, and P(confident), which represents the percentage of residues that were confidently predicted by AlphaFold2 with pLDDT greater than 70. Boxplot (n = 555) is plotted with whiskers extending to ±1.5 × IQR. c Two examples of potentially well-folded de novo gene candidates: CG42590 and CG43070. In both cases, the core regions were predicted with very high confidence (blue). d The predicted structural models of the 19 potentially well-folded de novo genes retained highly similar structural folds during MD simulations with pairwise TM-score of representative structures close to or larger than 0.70. Source data is provided in the source data file.