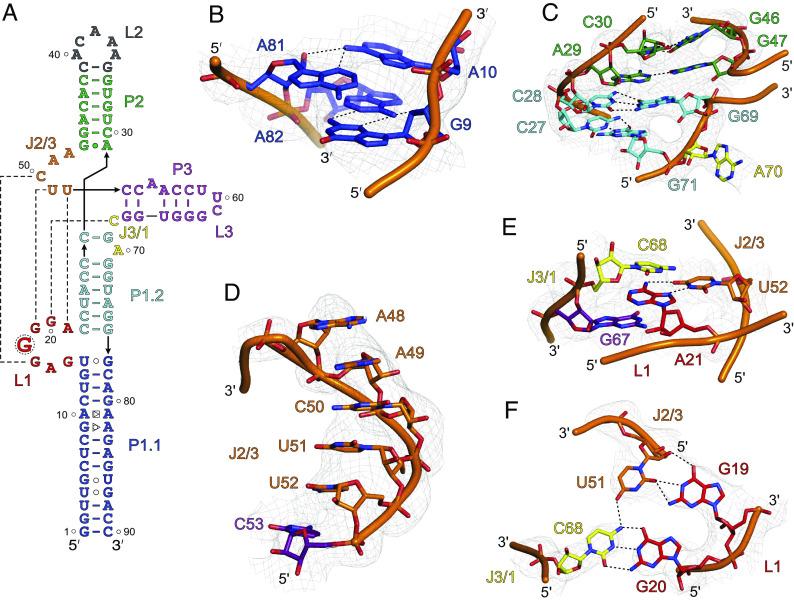
Fig. 3.
The unique features of SCV PTE crystal structure. (A) The crystal-derived secondary structure of the PTE RNA. The dotted lines represent tertiary interactions between the nucleotides (B) The non-canonical G9•A82 (sugar-edge-Hoogsteen) and A10•A81 (Hoogsteen-sugar-edge) pairs within the P1.1 helix. (C) The structure and helical stacking interactions within the J1/2. (D) Helical base-stacking of the J2/3 nucleotides that interact with L1 nucleotides to form a pseudoknot structure. (E) Additional interactions between the J2/3. The U52 base pairs with the A21, which sandwiches between the cross-strand stacking of the G67 and C68. (F) The J3/1 C68 nucleotide forms a canonical base pair with G20 and contacts U51 through a hydrogen bond that creates a planer tetrad structure involving G19 and G20, U51, and C68. Gray mesh represents the composite simulated anneal-omit 2|Fo|-|Fc| electron density map at contour level 1σ and carve radius 2.5 Å, and dotted black lines depict the heteroatoms within hydrogen bonding distances (≤3.0 Å).