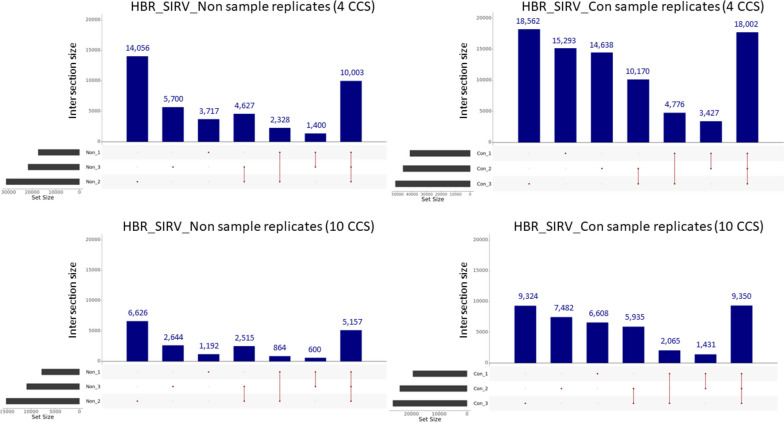
Fig. 6.
Detected isoforms supported by at least four or ten Circular Consensus Sequencing “CCS” reads between sample prep methods. The samples include Human Brain Reference RNA mixed with Spike-In RNA Variants “SIRV” prepared with the PacBio Full-Length Isoform Concatemer Sequencing workflow “HBR_SIRV_Con” or prepared with a non-concatemer Iso-Seq workflow “HBR_SIRV_Non”. Upset plot illustrating unique isoforms identified by GFFCompare analysis for the HBR_SIRV_Non and HBR_SIRV_Con sample set with required isoform read support of at least four or ten CCS reads. Dots indicate isoforms unique to the sample, and a line connecting two or more dots indicate isoforms detected in multiple sample replicates. The numbers listed above the bar graphs represent the number of unique isoforms detected for that sample(s)