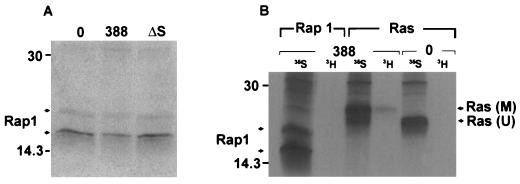
FIG. 6.
Investigation of Rap1 modification. (A) Examination of Rap1 mobility in HT29 cells following exposure to 388 or ΔS bacteria. [35S]methionine-labeled cells were prepared and cocultured with the indicated bacterial strains as described for Fig. 1B. Rap1 proteins were immunoprecipitated with rabbit polyclonal Rap1–Krev-1 (121) antibody and detected by SDS-PAGE, followed by fluorography. Arrows indicate 20- and 24-kDa Rap1 proteins. (B) Comparison of ADP-ribosylation of Ras and Rap1 in HT29 cells exposed to strain 388. Cellular proteins were labeled with [35S]methionine (35S), or intracellular NAD pools were labeled by treating HT29 monolayers with 5 μg of actinomycin D/ml to reduce RNA synthesis and then radiolabeling with [3H]adenosine (3H) for 18 h. Cells were either left untreated (0) or exposed to 108 CFU of strain 388 bacteria/ml. Ras and Rap1 were immunoprecipitated and subjected to SDS-PAGE and fluorography as described above. M, modified Ras; U, unmodified Ras. Molecular masses (in kilodaltons) are indicated.