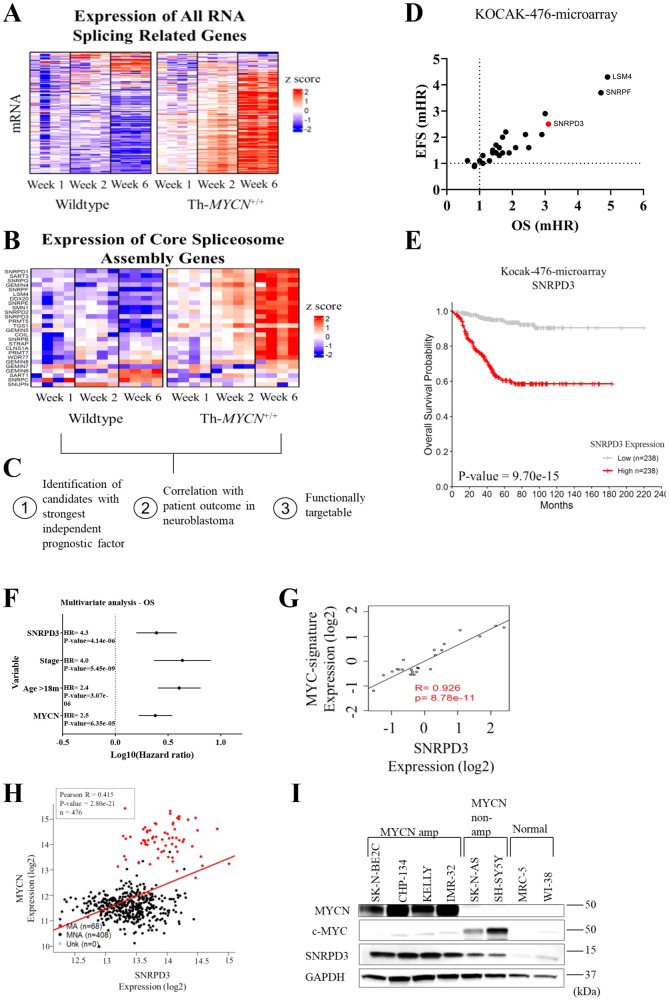
Fig. 1. Some core snRNP assembly genes are up-regulated in neuroblastoma and are prognostic for patient outcomes.
A heat map of a mRNA microarray analysis on ganglia cells collected from transgenic Th-MYCN + /+ and wildtype mice at 1, 2, and 6 weeks of age assessing the expression of (A) all RNA splicing-related genes, or (B) the core spliceosome assembly genes. C All 26 core spliceosome assembly genes (as defined by the GO term, GO:0000387) were assessed based on the three given criteria to identify possible candidate genes. D Overall (OS) and event-free survival (EFS) multivariate CoxPH analysis using the Kocak (n = 476) neuroblastoma patient cohort was conducted on the core spliceosome assembly genes (as defined by the GO term, GO:0000387). The dot plot represents the median hazard ratios (mHR) from the CoxPH models for each gene regarding EFS and OS. E Kaplan–Meier OS curve obtained from Kocak neuroblastoma patient cohort (n = 476) dichotomised on median SNRPD3 expression. F OS multivariate CoxPH analysis on Kocak cohort (n = 649) with SNRPD3 gene expression against classic neuroblastoma prognostic factors, dots represent the median HR (hazard ratio) whilst lines represent the 95% confidence interval. G Scatter plot with a linear regression fit of mRNA expression levels from the Th-MYCN+/+ mouse tissues for MYC-signature vs SNRPD3 gene expression (6 weeks of age, microarray, log2). H Scatter plot with linear regression fit for the Kocak neuroblastoma patient cohort for MYCN vs SNRPD3 gene expression (mRNA microarray, log2 expression). I Western blot analysis for SNRPD3, cMYC and MYCN expression in a range of MYCN-amplified (SK-N-BE(2)-C, CHP-134, KELLY, IMR-32), MYCN non-amplified (SK-N-AS, SH-SY5Y), and normal lung fibroblasts (MRC-5, WI-38) cells. GAPDH is a protein loading control. Western blot is a representative image of n = 3 independent experiments.