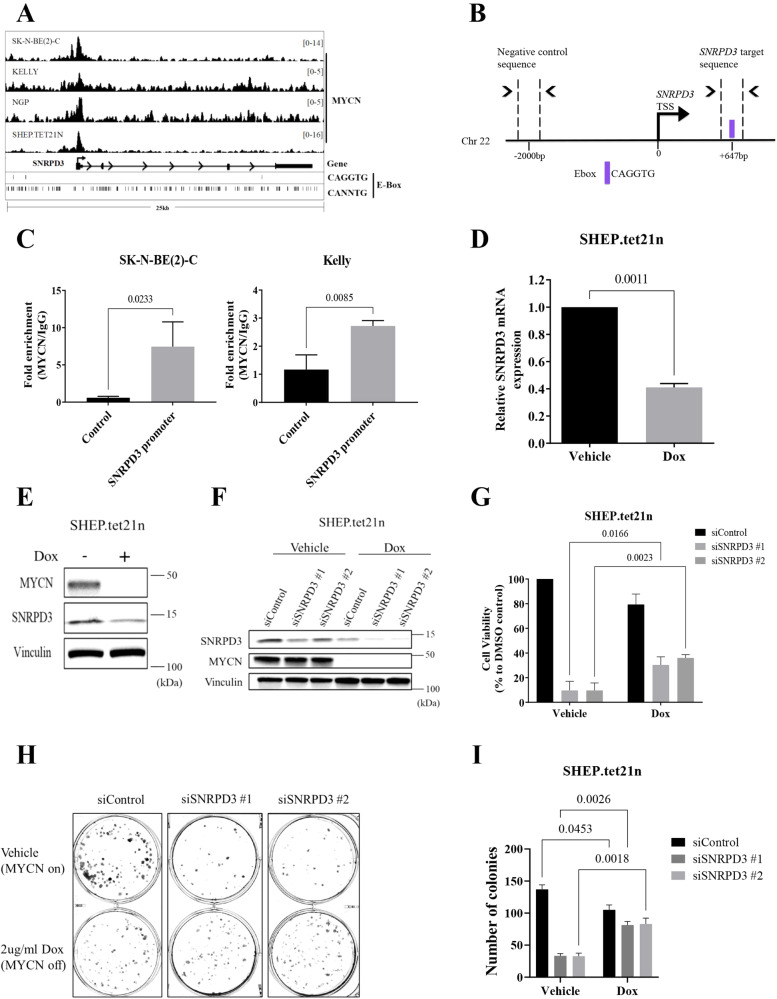
Fig. 2. MYCN regulates the expression of the SNRPD3 gene in neuroblastoma cells.
A ChIP-sequencing traces from publicly available databases [3] for MYCN binding the SNRPD3 gene in a panel of MYCN expressing neuroblastoma cells detailing MYCN binding motifs at canonical (CACGTG) and non-canonical (CANNTG) E-boxes B Schematic representation of the SNRPD3 target sequence (+647 bp from SNRPD3 TSS) and negative control target sequence (−2000 bp from SNRPD3 TSS) used for quantitative real time PCR (QPCR) following chromatin-immunoprecipitation (ChIP), detailing the MYCN peak summit and its distance from TSS. C ChIP-qPCR assay for the negative control region or the SNRPD3 promoter containing the MYCN binding site in SK-N-BE(2)-C and KELLY cells. Results represent n = 3 independent biological replicates, mean ± SEM. P-values were determined by two-tailed t test comparing control against SNRPD3 promoter. D SNRPD3 mRNA and E protein expression measured in SHEP.tet21n cells treated with doxycycline (Dox; MYCN off) or without Dox (Vehicle; MYCN on). Results represent n = 3 independent biological replicates, mean ± SEM, where P values were determined by two-tailed t test comparing vehicle against Dox. F A representative immunoblot of MYCN and SNRPD3 expression in SHEP.tet21n cells treated with either vehicle or Dox following SNRPD3 siRNA knockdown. G Cell viability was measured 72 h after SHEP.tet21n cells treated with either vehicle (DMSO) or Dox were transfected with control siRNA, or the two SNRPD3 siRNAs. Results represent n = 3 independent biological replicates, mean ± SEM, where the P value was determined through a two-way ANOVA, with Tukey’s multiple comparison tests. H SHEP.tet21n cells treated with either vehicle or Dox were transfected with control siRNA or the two SNRPD3 siRNAs for 10 days, followed by colony formation assays. Differences in colony formation were compared to siControl and I number of colonies quantified. Results represent n = 3 independent biological replicates, mean ± SEM, where the P value was determined through a two-way ANOVA, with Tukey’s multiple comparison tests.