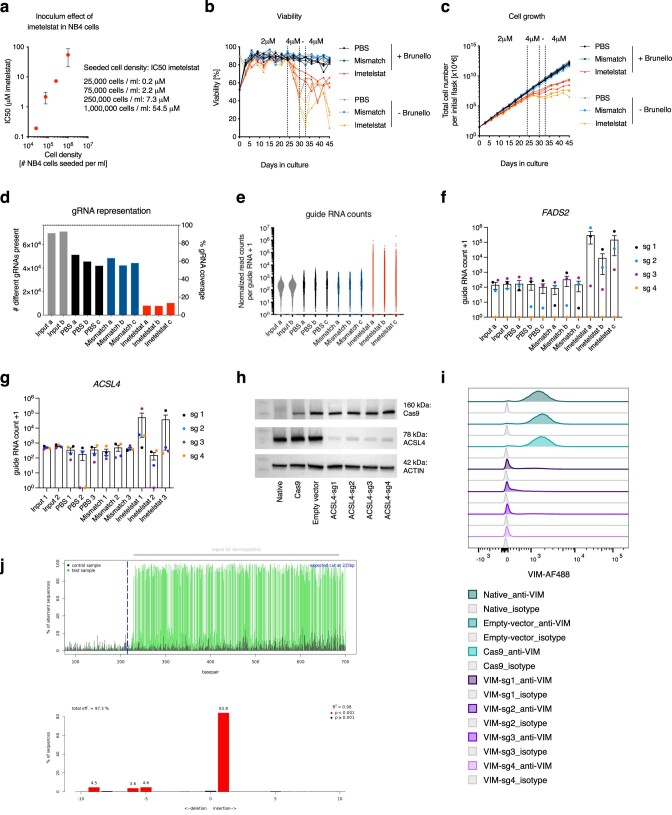
Extended Data Fig. 4. Identification of key mediators of imetelstat efficacy using a genome-wide CRISPR/Cas9 screen.
a, Identification of an imetelstat inoculum effect by quantification of IC50 values in NB4 cultures seeded at different densities: IC50 = 0.2 microM (25,000 cells per ml); IC50 = 1.5 microM (75,000 cells per ml); IC50 = 7.2 microM (250,000 cells / ml); IC50 = 31.3 microM (1,000,000 cells / ml). Dots represent mean IC50 values ± SEM from n = 2 independent experiments. b,c, Viability (b) and cell growth analysis (c) of Brunello-library-transduced NB4 cells cultured in the presence of vehicle control (PBS), mismatch control (MM1), or imetelstat compared to non-transduced NB4 cells over a time course of 45 days. The respective concentrations are indicated in the panel. N = 3 independent biological replicates per genotype and treatment condition. d,e, Next generation sequencing of DNA isolated from Brunello library-transduced NB4 cells harvested at day 45 in culture treated with vehicle control (PBS; black bars), mismatch control (MM1; blue bars), or imetelstat (red bars), and compared to DNA isolated from Brunello library-transduced NB4 cells before treatment as input control (gray bars): d, The number of different guide RNAs present (left y-axis) and percentage of guide RNA coverage (right y-axis); e, the read counts obtained from each guide RNA. f,g, Read counts obtained from n = 4 independent FADS2 (f) or n = 4 independent ACSL4 (g) targeting guide RNAs in the input, vehicle (PBS), mismatch (MM1) or imetelstat-treated Brunello-transduced NB4 cultures. Data are presented as mean ± SEM. h-j, Confirmation of efficient CRISPR/Cas9 - generated knockdowns in human AML cell lines by ACSL4 western blotting (h; additional biological replicates are provided as supplement), flow cytometric analysis of intracellular VIM expression (i), and FADS2 gene editing by tracking of indels by decomposition analysis using TIDE analysis tool (http://shinyapps.datacurators.nl/tide/) (j).