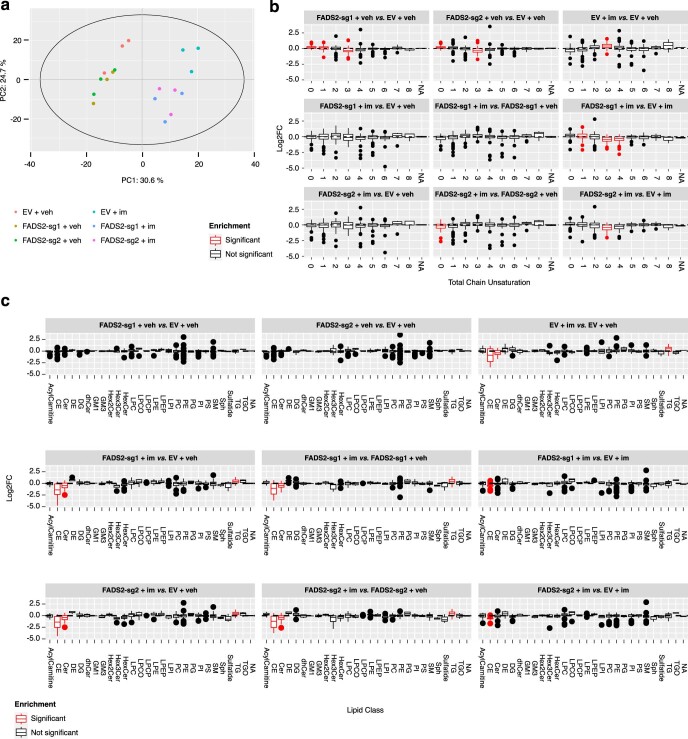
Extended Data Fig. 5. Lipidomics analysis of imetelstat-treated FADS2-edited or non-edited NB4 cells.
Targeted lipidomics analysis on 593 lipid species and their desaturation levels. a, PCA plot on normalized peak areas of lipid species in FADS2-edited (FADS2-sg1, FADS2-sg2) or non-edited (empty-vector control) NB4 cultures supplemented for 24h with either 4 microM imetelstat or vehicle control. b,c, Differential enrichment analysis using lipidR tool of unsaturated bonds in all measured lipid species (b), and lipid species according to lipid classes (c). Red box plots denote significantly different comparisons with P < 5 × 10−3. N = 3 independent replicates per condition from distinct cell passages. Lipid classes: Acylcarnitines (AcylCarnitine), Cholesteryl ester (CE), Ceramide (Cer), Desmosterol (DE), Diacylglycerol (DG), Dihydroceramide (dhCer), GM1 ganglioside (GM1), GM3 ganglioside (GM3), Dihexosylceramide (Hex2Cer), Trihexosylcermide (Hex3Cer), Monohexosylceramide (HexCer), Lysophosphatidylcholine (LPC), Lysoalkylphosphatidylcholine (LPCO), Lysoalkenylphosphatidylcholine (LPCP), Lysophosphatidylethanolamine (LPE), Lysoalkenylphosphatidylethanolamine (LPEP), Lysophosphatidylinositol (LPI), Phosphatidylcholine (PC), Phosphatidylethanolamine (PE), Phosphatidylglycerol (PG), Phosphatidylinositol (PI), Phosphatidylserine (PS), Sphingomyelin (SM), Sphingosine (Sph), Sulfatide (Sulfatide), Triacylglycerol (TG), Alkyldiacylglycerol (TGO). NA: Molecules which could not be parsed by lipidr (for example ubiquinone, free cholesterol).