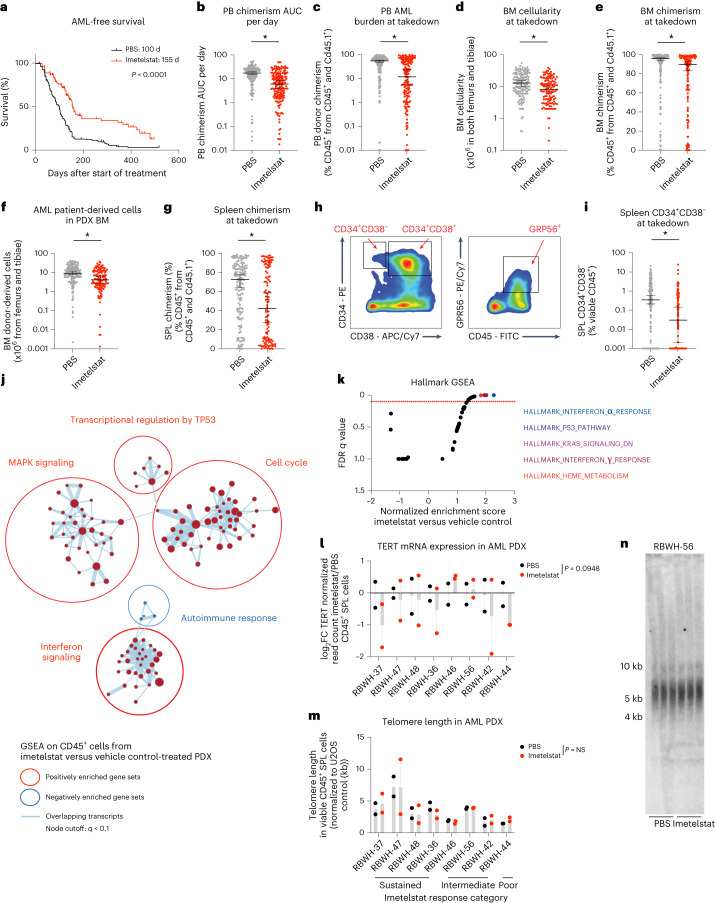
Fig. 2. The efficacy of imetelstat in a randomized phase II-like preclinical trial in AML PDX.
a, Two-tailed Kaplan–Meier survival analysis of vehicle control (PBS; n = 180) or imetelstat-treated (n = 180) AML PDX. P < 1 × 10−4 according to Gehan–Breslow–Wilcoxon. b–g, Analysis of AML disease parameters. Peripheral blood (PB) donor chimerism area under the curve (AUC) per day (b), end point PB donor chimerism (c), bone-marrow (BM) cellularity (d), BM chimerism (e), the number of AML donor-derived cells in PDX BM (f) and splenic (SPL) donor chimerism (g). h,i, Flow cytometric analysis of AML surface marker expression CD34, CD38 and GPR56. Gating strategy (h). The percentage of CD34+CD38− viable CD45+ SPL singlets (i). Data are presented as median ± 95% confidence interval (CI) (b–g,i). Statistical analysis was performed on log-transformed data using an unpaired two-sided t-test, considering detection limits at 1 × 10−3. P = 2.21 × 10−10 (b), P = 7.79 × 10−8 (c), P = 1.37 × 10−4 (d), P = 7.32 × 10−3 (e), P = 8.82 × 10−5 (f), P = 1.83 × 10−3 (g), P = 7.44 × 10−5 (i). Asterisks (*) denote statistically significant comparisons with P < 5 × 10−2. j,k, GSEA on RNA-seq data from sorted viable hCD45+ cells collected from imetelstat or PBS-treated AML PDXs. n = 16 AML PDXs per treatment group. Cytoscape nodes represent gene sets with a cutoff of q < 0.1 (j); GSEA on hallmark signatures with the top five enriched signatures highlighted in color (k). l–n, TERT messenger RNA (mRNA) expression results obtained from RNA-seq analysis described as above (l). FC, fold change. Telomere length in viable CD45+ SPL cells from imetelstat versus PBS-treated AML PDXs measured by qPCR (m) and confirmed by telomeric restriction fragment analysis (n). Statistical analysis (l,m) was based on paired two-tailed t-tests comparing AML PDXs treated with imetelstat (n = 16) or PBS (n = 16). P = 9.48 × 10−2 (l), P > 5 × 10−2 (m). Data are presented as mean ± s.e.m. NS, not significant.