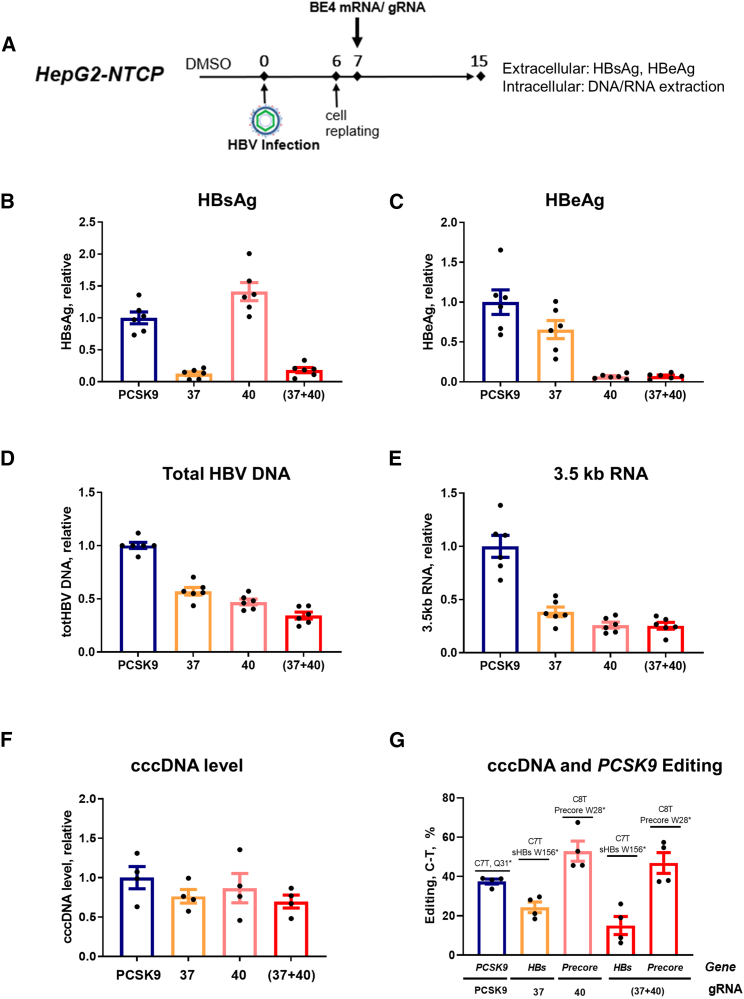
Figure 2.
Effect of CBE and gRNAs g37, g40, or their combination on HBV extracellular and intracellular parameters in HepG2-NTCP cells
(A) Experimental scheme. A protocol similar to Figure S1B was used with a few modifications. All of the samples were collected at 15 dpi. (B and C) Extracellular HBsAg and HBeAg were measured by ELISA. (D) Total HBV DNA was quantified by quantitative (qPCR) from DNA extracted from cell lysates. (E) Total cellular RNA was extracted and HBV 3.5-kb RNA levels were quantified by quantitative reverse transcription-PCR (qRT-PCR). Data were normalized to PCSK9 control gRNA targeting Proprotein convertase subtilisin/kexin type 9 (gene unrelated to HBV). (F) cccDNA level was assessed by qPCR on the DNA samples pretreated with ExoI/III in HepG2-NTCP. (G) Level of the C>T functional editing that leads to the introduction of the stop codons in HBs and Precore genes, assessed by NGS on ExoI/III-treated cccDNA samples from HepG2-NTCP, as well as PCSK9 (assessed on total DNA). Data are represented as mean ± SEM for n = 4 to 6 replicates.