Abstract
Interferon regulatory factor 4 (IRF4) is a transcription factor (TF) and key regulator of immune cell development and function. We report a recurrent heterozygous mutation in IRF4, p.T95R, causing an autosomal dominant combined immunodeficiency (CID) in seven patients from six unrelated families. The patients exhibited profound susceptibility to opportunistic infections, notably Pneumocystis jirovecii, and presented with agammaglobulinemia. Patients’ B cells showed impaired maturation, decreased immunoglobulin isotype switching, and defective plasma cell differentiation, whereas their T cells contained reduced TH17 and TFH populations and exhibited decreased cytokine production. A knock-in mouse model of heterozygous T95R showed a severe defect in antibody production both at the steady state and after immunization with different types of antigens, consistent with the CID observed in these patients. The IRF4T95R variant maps to the TF’s DNA binding domain, alters its canonical DNA binding specificities, and results in a simultaneous multimorphic combination of loss, gain, and new functions for IRF4. IRF4T95R behaved as a gain-of-function hypermorph by binding to DNA with higher affinity than IRF4WT. Despite this increased affinity for DNA, the transcriptional activity on IRF4 canonical genes was reduced, showcasing a hypomorphic activity of IRF4T95R. Simultaneously, IRF4T95R functions as a neomorph by binding to noncanonical DNA sites to alter the gene expression profile, including the transcription of genes exclusively induced by IRF4T95R but not by IRF4WT. This previously undescribed multimorphic IRF4 pathophysiology disrupts normal lymphocyte biology, causing human disease.
INTRODUCTION
Inborn errors of immunity (IEI) are a heterogeneous group of gene defects characterized by increased susceptibility to infections, autoimmunity, autoinflammation, allergy, and, in some cases, cancer. During recent years and after the advent of high-throughput sequencing, the rate of discovery of new genetic lesions causing IEI has skyrocketed (1). Yet a large percentage of patients with an IEI lack a genetic diagnosis. Furthermore, because of the complexity of designing gene-tailored experiments to assess the pathogenicity of genetic variants and the scarcity of patients with similar genetic diseases, the molecular consequences of many IEI-causing mutations are poorly understood (2).
The human interferon (IFN) regulatory factor (IRF) family is a group of nine transcription factors (TFs) that regulate critical innate and adaptive immune processes (3). The IRF family is typically characterized by the ability to recognize promoters containing the IRF consensus sequence (GAAA) (4). IRF4 is a predominantly hematopoietic TF that is pivotal for the development and function of B and T cells as well as dendritic cells and macrophages (5-7). IRF4 binds to the canonical IFN-stimulated response elements (ISREs) as a homodimer, and it engages erythroblast transformation-specific IRF composite elements (EICEs) and activating protein 1 (AP-1)–IRF composite elements (AICE1 or AICE2) as a heterodimer requiring PU.1 or SPIB, or AP-1 family members, respectively, for high-affinity interactions (5, 8, 9). Irf4−/− mice have severely impaired B and T cell activation and differentiation, along with a profound reduction in serum immunoglobulins (Ig) of all isotypes (3).
Here, we describe a human immune disorder identified in seven patients from six unrelated families across four continents. This newly described form of combined immunodeficiency (CID) is caused by the same heterozygous germline mutation in the DNA binding domain (DBD) of IRF4 (p.T95R), and mouse knock-in models accurately recapitulated the human phenotype. Particularly notable was the mechanism of this human disease with a simultaneous multimorphic combination of dominant loss, gain, and new functions for IRF4.
RESULTS
Heterozygous T95R IRF4 variant identified in seven patients with profound CID
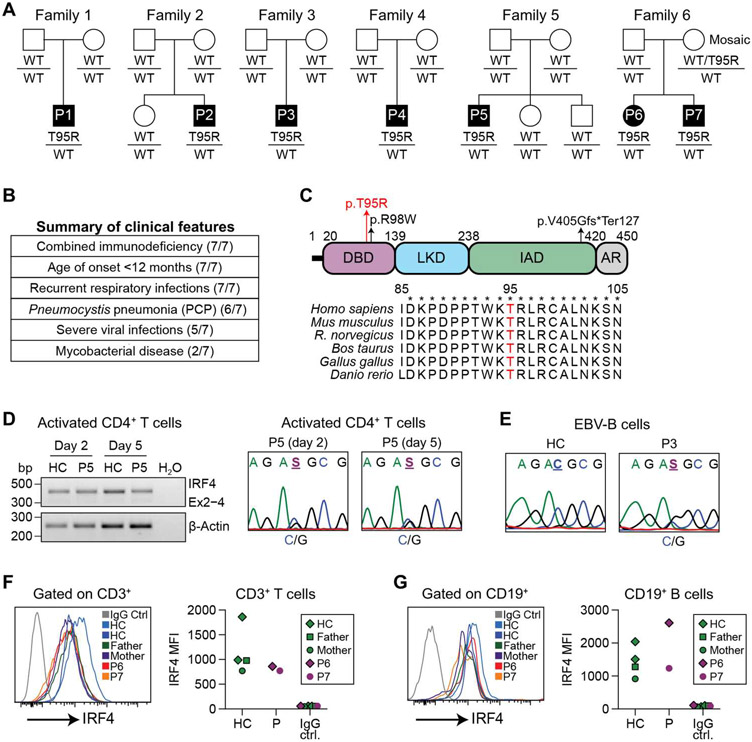
We investigated seven patients with profound CID from six kindreds of diverse ethnic origins (Fig. 1A). All affected individuals suffered with early onset (<1 year of age) recurrent sinopulmonary infections, with the opportunistic pathogen Pneumocystis jirovecii causing pneumonia in most individuals. Other clinical features indicative of their substantial immune compromise were severe viral infections [notably cytomegalovirus (CMV) and Epstein-Barr virus (EBV)], localized infection with weakly pathogenic [i.e., vaccine-strain Bacille Calmette-Guerin (BCG)] or pathogenic mycobacteria, (i.e., Mycobacterium bovis), and chronic diarrhea (Fig. 1B and table S1; patient summaries in Supplementary Materials and Methods). Next-generation sequencing was performed on all patients, with Sanger sequencing and familial segregation used for confirmation (fig. S1, A and B). All seven individuals carried the same heterozygous variant in the gene IRF4 (IRF4 NM_001195286.2 c.284C>G, p.T95R; henceforth designated as IRF4T95R) (Fig. 1C). This variant was de novo in all patients, except in the case of kindred six (P6 and P7), where the mother was found to be mosaic with 4 of 124 reads mapping to the variant in peripheral blood genetic testing (fig. S1B). The IRF4T95R variant has not been reported in population databases of controls (fig. S1C) and is predicted to be pathogenic by a variety of in silico pathogenicity models (table S2). IRF4T95R has been reported in the Catalogue of Somatic Mutations in Cancer associated with adult T cell leukemia (10).
Fig. 1. Identification of a unique heterozygous mutation in the IRF4 gene in seven CID patients.
(A) Pedigrees of seven patients from six unrelated families with an identical IRF4 mutation (c.284C>G, p.T95R). Affected individuals are indicated by the filled symbols. (B) Summary of the major clinical features in these patients. (C) Schematic representation of the IRF4 protein (isoform 2; National Center for Biotechnology Information accession#: NP_001182215.1). LKD, linker domain; IAD, IRF association domain; AR, autoregulatory region. The T95R substitution is indicated by a red arrow. A recently identified heterozygous loss-of-function (LOF) mutation (c.292C>T, R98W) associated with Whipple’s disease and a homozygous splicing mutation (c.1213-2A>G, p.V405Gfs*Ter127) causing human CID are also indicated. Bottom: alignment of the amino acids in the DBD of IRF4 from different species. T95 is shown in red. (D) Left: CD4+-naïve T cells from P5 were stimulated with anti-CD3/CD28 beads, IL12, and Activin A for 2 and 5 days. Total RNAwas isolated from 500 cells and analyzed by reverse transcription PCR (RT-PCR). Right: Sanger sequencing of RT-PCR products. HC, healthy controls. (E) Sanger sequencing of RT-PCR products of EBV B cells from P3. (F and G) Flow cytometric analysis (intracellular staining) for IRF4 protein expression in gated CD3+ T cells (F) or CD19+ B cells (G) Left: representative profiles. Right: mean fluorescence intensity (MFI) of IRF4 protein from P6 and P7, their parents, and two HC.
IRF4 mRNA and protein is expressed normally in IRF4T95R patient lymphocytes, but broad immune architecture is disrupted
Because IRF4 is highly expressed in lymphocytes, we investigated how the IRF4T95R variant affects the total IRF4 mRNA and protein expression in these cells. We found that total IRF4 mRNA expression was comparable with controls in activated CD4+ T cells (Fig. 1D, left). Sanger sequencing of cDNAs extracted from both activated CD4+ T cells (Fig. 1D, right) and EBV-immortalized B (EBV-B) cells (Fig. 1E) suggested that the wild-type (WT) and mutant alleles are likely expressed at similar levels. We also found that IRF4 protein levels of the patients were comparable to those of controls in unstimulated CD3+ T cells (Fig. 1F), CD19+ B cells (Fig. 1G), expanded T cell blasts (fig. S1D, left), activated CD4+ T cells (fig. S1D, right), and EBV-B cells (fig. S1E). In combination, these results show that the T95R mutation does not affect the total IRF4 mRNA and protein levels.
The infection pattern experienced by the individuals carrying IRF4T95R was not consistent with a defect in just one immune compartment, suggesting a broad immunological phenotype. To define the overall immune architecture caused by IRF4T95R in an unbiased manner, we applied cytometry by time of flight (CyTOF). Combining unsupervised analysis with manual clustering, we found that the distribution of B cells and CD4+ and CD8+ T cells was different between healthy controls and patients (fig. S2, A to C). These data suggested that IRF4T95R causes a broad immunological phenotype, most notably affecting B and T cells, that we set out to systematically decipher.
IRF4T95R patients have multiple defects in B cell development and function
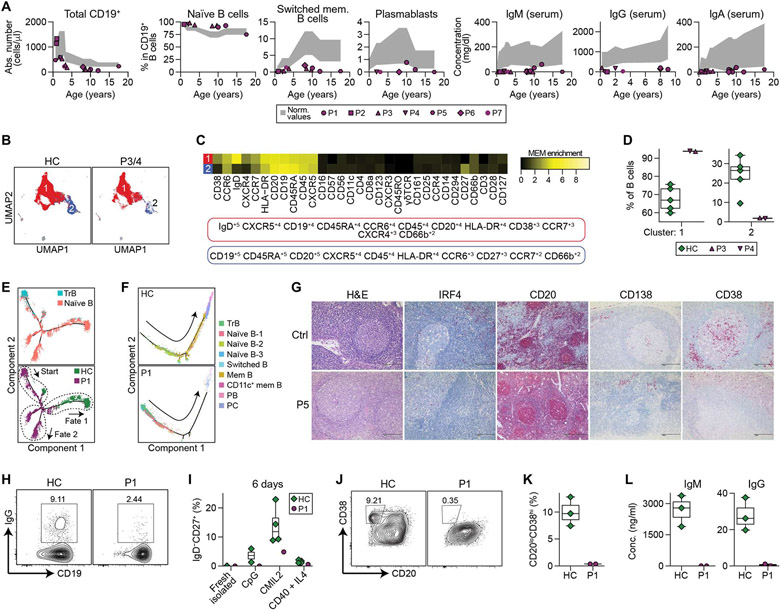
IRF4 is critical for B cell development and differentiation (7, 11). All patients were virtually agammaglobulinemic with severely reduced serum concentrations of IgM, IgG, and IgA (Fig. 2A and tables S3 to S9). B cell enumeration revealed that all patients had reduced but not absent total CD19+ B cells, with a developmental arrest characterized by increased naïve B cells, reduced class-switched memory B cells, and decreased plasmablasts (Fig. 2A and tables S3 to S9). The B cell differentiation defect was further refined in high-dimensional space using CyTOF (Fig. 2, B to D, and fig. S2, A to C), flow cytometric analysis (fig. S2, D and E), and single-cell RNA sequencing (scRNA-seq) on sorted CD19+ B cells (Fig. 2, E and F). Pseudotime inference indicated that IRF4T95R transitional B (TrB) cells matured toward entirely different fates than those from a healthy control (Fig. 2E) and further confirmed an accumulation of TrB and naïve B cells with few terminally differentiated B cell subsets (i.e., memory B and plasma cells) in IRF4T95R B cells (Fig. 2F). Moving beyond peripheral blood analysis, we confirmed that IRF4T95R patient bone marrow (fig. S2F) and lymph node (Fig. 2G) also had scant CD38+ or CD138+ plasmablasts.
Fig. 2. Impaired Ig gene CSR and defective memory B and plasma cell differentiation in IRF4T95R patients.
(A) Total number of B cells, proportions of naïve B, switched memory B and plasmablasts among CD19+ B cells and serum Ig levels in the seven patients. Age-matched reference ranges are shown in gray. Detailed data are shown in tables S3 to S9. (B to D) CyTOF analysis of the B cell cluster from fig. S2 (A to C) performed in P3, P4, and five HC. (B) Dimensional reduction by uniform manifold approximation and projection (UMAP) of the two major populations obtained by unsupervised clustering using FlowSOM. (C) MEM heatmap and tags showing the markers that characterize each population. (D) Proportions of clusters 1 and 2 among total B cells from fig. S2 (A to C). (E and F) Pseudotime analysis based on scRNA-seq of purified peripheral blood B cells performed in an HC and P1. (E) Pseudotime analysis of transitional and naïve B cell subpopulations. TrB, transitional B. (F) Pseudotime analysis of B cell differentiation into memory B and plasma cells. (G) Immunohistochemical staining of lymph nodes of P5 and an unaffected control (Ctrl). H&E, hematoxylin and eosin. PB, plasmablast. PC, plasma cell. (H) Purified naïve B cells of an age-matched HC and P1 were cultured with CD40L plus IL4 for 6 days and analyzed for the percentages of IgG+ cells by flow cytometry. (I) Purified naïve B cells of four HC and P1 were stimulated with CpG ODN 2006 alone, CpG + F(ab′)2 anti-IgM and IL2 (CMIL2), or CD40L + IL4 for 6 days and analyzed for the generation of CD19+IgD−CD27+ memory B cells. (J to L) Purified naïve B cells of HC and P1 were stimulated with CMIL2 for 6 days and analyzed for the induction of CD20−CD38+ plasmablasts and Ig secretion. (J) Representative FACS profiles. (K) Results of three HC and two experiments of P1. (L) IgM and IgG levels in the culture supernatants.
Having demonstrated a marked impairment in B cell differentiation in patient peripheral blood using multiple complementary technologies (Fig. 2, A to D, and fig. S2, A to E), we designed experiments to determine whether this defect was intrinsic or extrinsic to the IRF4T95R B cells. To test this, we isolated naïve B cells from patients and controls and exposed them to classic B cell stimuli. When compared with healthy controls, purified naïve IRF4T95R B cells or total peripheral blood mononuclear cells (PBMCs) generated fewer class-switched (Fig. 2H and fig. S2G) and memory B cells (Fig. 2I), and the induction of plasmablasts (Fig. 2, J and K, and fig. S2, H and J) and Ig secretion was lost or greatly reduced (Fig. 2L and fig. S2, I and K). Reduced B cell differentiation was not due to impaired cell proliferation (fig. S2L). Collectively, these data demonstrate that IRF4T95R B cells have an intrinsic defect in differentiation into memory B cells, plasmablasts and plasma cells, as well as Ig secretion.
IRF4T95R patient T cell differentiation is largely intact, but effector function is abnormal
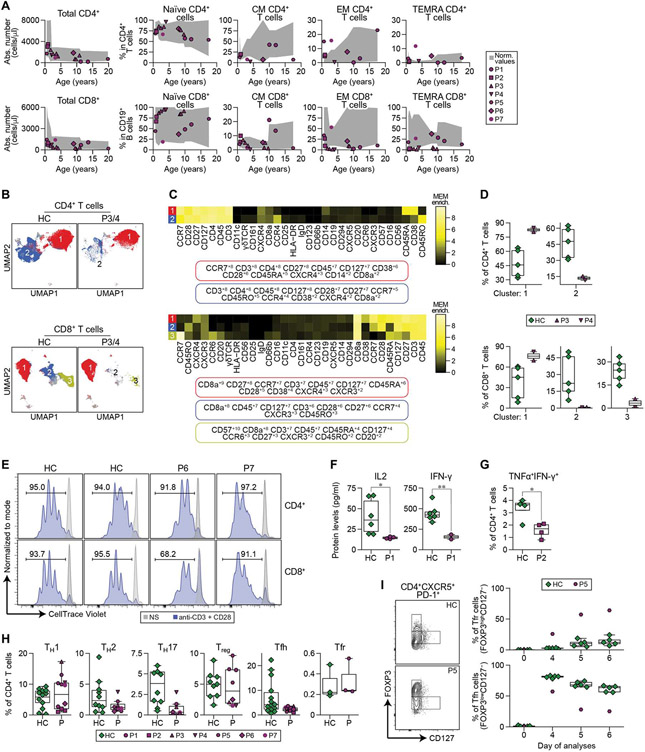
In contrast to the notable impact on B cell differentiation and biology, IRF4T95R patients had relatively normal numbers of total T cells in peripheral blood (Fig. 3A). Appreciating that IRF4 has been linked to both CD4+ and CD8+ T cell effector function and that both P. jirovecii infection and mycobacterial disease are not commonly found in patients with isolated B cell deficiencies (12), we proceeded with higher-resolution phenotyping and functional interrogation. The proportions of naïve, central memory (CM), effector memory (EM), and EM re-expressing CD45RA (EMRA or TEMRA) T cells among the CD4+ population were relatively normal (Fig. 3A, top panels), whereas CD8+ CM, EM, and TEMRA cells were reduced in some patients (Fig. 3A, bottom panels). In multidimensional space, both CyTOF and scRNA-seq revealed an abnormal distribution of CD4+ and CD8+ T cells (Fig. 3, B to D, and figs. S3, A and B and S4 and S5). Further functional studies confirmed that in IRF4T95R T cells, T cell receptor Vβ (TCRVβ) usage was not affected (fig. S3C), and T cells were activated and proliferated normally upon TCR (i.e., anti-CD3/CD28) stimulation (fig. S3, D and E). Cytokine production is another critical effector function of T cells. Stimulated IRF4T95R T cells produced less interleukin-2 (IL2) and IFN-γ and had less polyfunctional cells that produced both tumor necrosis factor–α (TNFα) and IFN-γ (Fig. 3, F and G). T helper (TH) cell commitment, evaluated by flow cytometry and CyTOF, showed that TH1, TH2, regulatory T (Treg), and T follicular regulatory (Tfr) cells were similar to healthy controls (Fig. 3H and table S10). In contrast, TH17 and T follicular helper (TFH) cells were reduced in most patients (Fig. 3H and table S10), and in vitro differentiation to TFH cells was reduced (Fig. 3, I and J). When considered in the context of the patients’ infectious history, these data indicate that IRF4T95R T cells have a defect in subset differentiation and function, although this defect is more subtle than that observed in B cells.
Fig. 3. Altered T cell differentiation and reduced T cell activation and cytokine production in IRF4T95R patients.
(A) Total number of CD4+ (top left) and CD8+ (bottom left) T cells and percentages of naïve, CM, EM, and EM re-expressing CD45RA (TEMRA) cells among CD4+ (top) and CD8+ (bottom) T cells. (B to D) CyTOF analysis of the CD4+ and CD8+ cell clusters in P3, P4, and five HCs from fig. S2 (A to C). (B) Dimensional reduction by UMAP showing the two major clusters in the CD4+ population and the three major clusters of the CD8+ population obtained by unsupervised clustering using FlowSOM. (C) MEM heatmap and tags showing the markers that characterize each population. (D) Proportions of the different clusters among CD4+ (top) and CD8+ (bottom) cells from fig. S2 (A to C). (E) PBMCs from two HC and P6 and P7 were labeled with CellTrace Violet and stimulated with anti-CD3/anti-CD28 dynabeads for 4 days. Top: gated CD4+T cells. Bottom: gated CD8+T cells. NS, nonstimulation. (F) PBMCs from HC and P1 were stimulated with anti-CD3 and anti-CD28 for 24 hours and analyzed for the amount of IL2 and IFN-γ in the culture supernatants. Results of six HC and two to four independent experiments of P1 are shown. (G) Intracellular staining of TNFα and IFN-γ in CD4+ T cells from a HC and P2 before and after phorbol 12-myristate 13-acetate + ionomycin stimulation. Mean ± SD of four experiments was shown. (H) TH cell subset distributions in T95R patients and HC. The proportions of TH1, TH2, TH17, Treg, TFH, and Tfr among peripheral CD4+T cells were determined by flow cytometric analysis for P1, P2, and P5 to P7 and by CyTOF for P3 and P4 (table S10). (I) Purified naïve T cells (CD3+CD4+CD45RA+) of P5 and five HC were subjected to a TFH/Tfr differentiation assay and were analyzed by flow cytometry on days 0, 4, 5, and 6. Left: representative FACS profiles on day 6. Right: Proportions of Tfr-like (FOXP3highCD127−) and TFH-like (FOXP3lowCD127+/−) cells among the CD4+PD-1+CXCR5+T cells were quantified. Red dots show means of three biological independent P5 replicates. Bars represent mean percentages of HC. Statistical significance was assessed by one-tailed Welch’s t test (F and G). *P < 0.05 and **P < 0.01.
IRF4T95R patient monocyte and NK cell evaluation showed no gross abnormalities
Monocyte and natural killer (NK) cells were evaluated as part of a comprehensive description of a unique IEI. Frequencies of classical, intermediate, and nonclassical monocytes were within normal ranges in patients analyzed (fig. S3E and table S11). CD107a degranulation studies of NK (fig. S3F) and CD8+ T cells (fig. S3G) were also within normal ranges in patients tested. Further and deeper studies will be required to rule out more nuanced defects in these cells.
Heterozygous p.T95R IRF4 knock-in mice recapitulate patient phenotype and provide additional insights into T95R pathophysiology
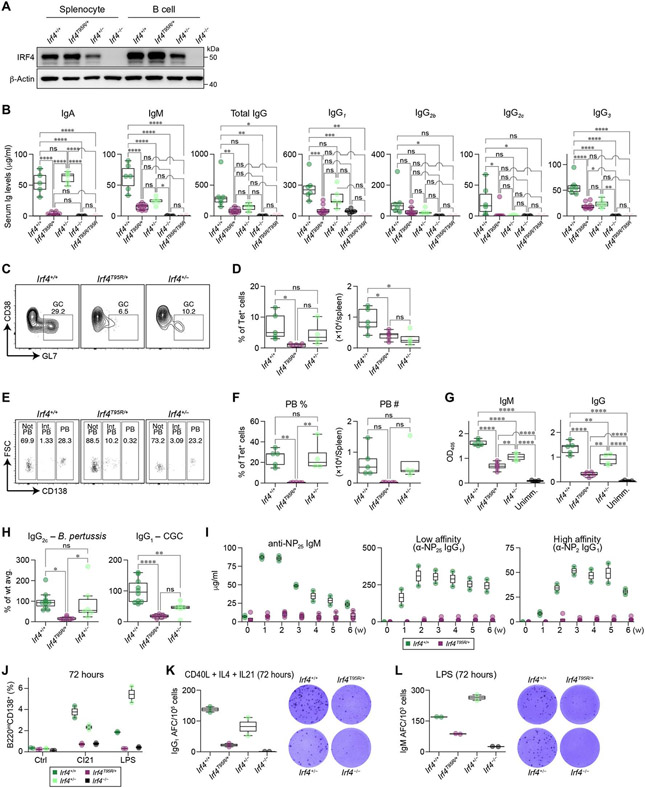
To obtain additional insights into the pathophysiological mechanisms of the disease caused by IRF4T95R, we generated knock-in mice carrying the same p.T95R variant in the heterozygous state (Ifr4T95R/+) and compared them with Itf4+/+, Irf4+/−, Irf4−/−, and Irf4T95R/T95R mice. Replicating the patient phenotype, Irf4T95R/+ splenocytes and B cells had IRF4 protein expression comparable to WT mice, whereas Irf4+/− had reduced protein expression and Irf4−/− were IRF4 protein deficient (Fig. 4A). Total numbers of B and T cells in the spleen (fig. S6A) and of B220+ B cells in the bone marrow and lymph nodes (fig. S6, B and C) were comparable among Irf4+/+, Irf4T95R/+, Irf4+/−, and Irf4−/− mice, although there was a moderate increase in B220+ cells in peripheral lymph nodes of Irf4T95R/+ relative to Irf4+/− mice (fig. S6C). Irf4−/− mice had barely detectable levels of serum Igs, whereas Irf4+/− mice had normal levels of serum IgA and total IgG but reduced IgM, IgG2c, and IgG3 levels (Fig. 4B). Notably, serum Ig levels were further reduced in Irf4T95R/+ mice compared with Irf4+/− mice and were very close to those found in Irf4−/− mice (Fig. 4B). Similar to Irf4−/− mice, T95R homozygous mice (Irf4T95R/T95R) had barely detectable levels of serum Igs (Fig. 4B). Total germinal center (GC) B cells were expanded in the spleens, mesenteric lymph nodes, and Peyer’s patches of Irf4T95R/+ mice at the steady state (fig. S6D) and in the spleen after immunization with sheep red blood cells (SRBC) (fig. S6E) or Plasmodium sporozoites (fig. S6F). Such expansions of GC B cells were not observed in Irf4−/− mice, consistent with previous studies (13-15), nor in Irf4T95R/T95R mice (fjg. S6D). In the Plasmodium sporozoite model, GC B cells specific for the immunodominant circumsporozoite protein (CSP) were decreased in Irf4T95R/+ mice compared with Irf4+/+ mice (Fig. 4, C and D), suggesting that there is an expansion of nonspecific GC B cells at the expense of antigen-specific GC B cells in Irf4T95R/+ mice. After Plasmodium immunization, Irf4T95R/+ mice generated few malaria CSP-specific CD138+ plasmablasts (Fig. 4, E and F) and had lower levels of CSP-specific IgM and IgG antibodies (Fig. 4G). Further defining the antibody production defect, IgG2c production after immunization with formalin-fixed Bordetella pertussis (TH1-dependent) was reduced in Irf4T95R/+ mice compared with Irf4+/+ and Irf4+/− mice (Fig. 4H, left), and IgG1 production against chicken γ-globulin (CGG) immunization (TH2-dependent) was also reduced in Irf4T95R/+ mice, although the difference between Irf4T95R/+ and Irf4+/− mice was not significant (Fig. 4H, right). After immunization of 4-hydroxy-3-nitrophenyl acetyl (NP)-conjugated to CGG (NP-CGG), the production of NP–specific IgM and both low- and high-affinity NP-specific IgG1 antibodies were barely detectable in Irf4T95R/+ mice compared with Irf4+/− mice (Fig. 4I). In line with earlier findings in Irf4−/− mice (16-18), Irf4T95R/+ mice also showed altered distribution of various T cell subsets (fig. S7, A to C).
Fig. 4. Heterozygous p.T95R IRF4 knock-in mice recapitulate the immunodeficiency phenotypes observed in IRF4T95R patients.
(A) WT and mutant IRF4 protein expression in total splenocytes and purified B cells from WT (Irf4+/+), Irf4T95R/+, Irf4+/−, and Irf4−/− male mice. (B) Serum Ig levels in male and female mice were measured by ELISA. (C) Irf4+/+, Irf4T95R/+, and Irf4+/− mice were immunized with Plasmodium sporozoites and analyzed for the proportions of GC B cells within the Tet+ CSP-specific B cells, ns, not significant. (D) Percentages of Tet+ (antigen-specific) cells within all GC B cells (left) and number (right) of Tet+ (antigen-specific) GC B cells (left). (E) Irf4T95R/+ mice were unable to generate CD138high PBs. FSC, forward scatter. (F) Percentages (left) and number (right) of Tet+ PBs in the spleen.(G) The production of CSP-specific IgM and IgG antibodies. Absorbance at 405 nm was measured, and the area under the curve was calculated in Prism 8 from the log (dilution) on the x axis and the absorbance at 405 nm on the y axis, fitting a sigmoidal curve. OD405, optical density at 405 nm. (H) Left: IgG2c production after immunization with formalin-fixed B. pertussis. Right: IgG1 production in response to CGG immunization. (I) WT and Irf4T95R/+ female mice were immunized intraperitoneally with 25 μg of NP-CGG in alum. Serum levels of NP-specific IgM and low and high-affinity IgG1 were determined each week by ELISA. (J to L) Naïve B cells purified from WT and mutant male mice were cultured for 72 hours in the presence of CI21 or LPS. The cells were then analyzed for the generation of CD138+ plasma cell by flow cytometry (J) and IgG1 and IgM secretion by ELISPOT (K and L). Each dot represents data from an individual mouse. AFC, antibody-forming cell. Statistical significance was determined by Tukey’s post hoc test (B, D, and F to H). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To clarify whether the impaired production of antibodies in Irf4T95R/+ mice was due to B cell–intrinsic defects in plasma cell differentiation, we cultured purified splenic B cells in medium alone or in the presence of CD40L + IL4 + IL21 (CI21) or lipopolysaccharide (LPS) for 72 hours. The percentage of B220inCD138+ cells in cultured Irf4T95R/+ B cells was lower than those in WT and Irf4+/− B cells and was indistinguishable from that in Irf4−/− B cells (Fig. 4J). In addition, the frequency of antibody-forming cells was reduced in Irf4T95R/+ B cells compared with WT or Irf4+/− B cells (Fig. 4, K and L). Further defining the B cell defect, we found that when splenic B cells from WT or Irf4T95R/+ Blimp1–green fluorescent protein (GFP) reporter mice were stimulated with LPS, Irf4T95R/+ B cells were able to activate Blimp1 transcription but failed to up-regulate CD138 expression (fig. S7, D and E), suggesting that Irf4T95R/+ B cells initiate, but fail to complete, plasma cell differentiation.
IRF4T95R B cells fail to undergo appropriate differentiation due to failure to activate IRF4 target genes critical for plasma cell differentiation
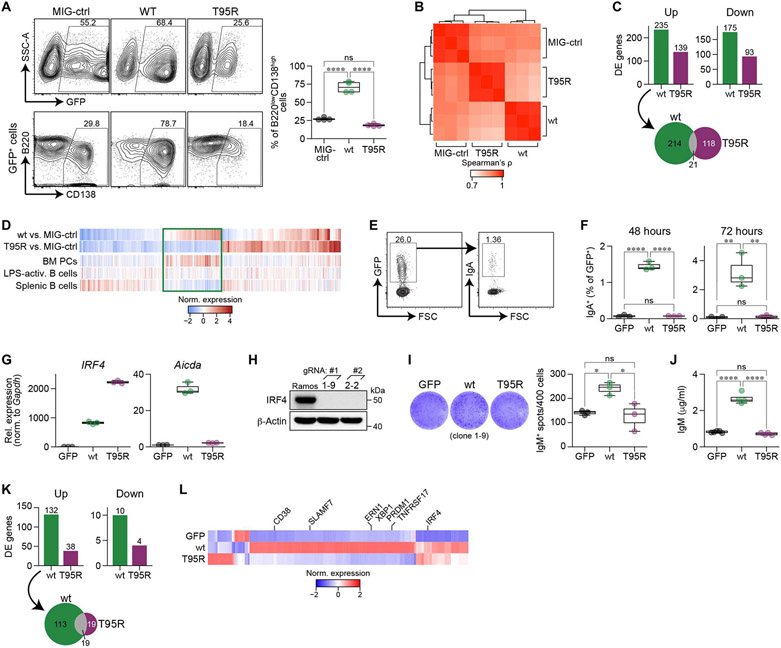
To understand why IRF4T95R B cells fail to appropriately differentiate into antibody-secreting plasma cells, we leveraged a variety of complementary model systems to assess different aspects of B cell development. First, mouse C57BL/6 splenic B cells were stimulated and transduced with IRF4WT, IRF4T95R, or control (MIG-ctrl) retroviruses (Fig. 5A and fig. S8A). Ectopic expression of IRF4WT robustly enhanced the formation of CD138high B220low plasmablasts compared with the MIG-ctrl (Fig. 5A). In contrast, IRF4T95R failed to enhance plasmablast formation (Fig. 5A). We next isolated IRF4WT-, IRF4T95R-, or MIG-ctrl–transduced B cells for RNA-seq analyses. Overall, IRF4T95R-transduced cells clustered separately from control or IRF4WT cells, indicative of a distinct gene expression program (Fig. 5B). Specifically, IRF4T95R regulated fewer (Fig. 5C, top) and unique (Fig. 5C, bottom) genes, with only a small overlap with genes differentially regulated by IRF4WT (Fig. 5C, fig. S8B, data file S1). Through comparison to other gene expression profiles, we found that IRF4T95R virtually lost the ability to regulate IRF4-driven and plasma cell–specific gene expression (Fig. 5D).
Fig. 5. IRF4T95R failed to induce plasma cell differentiation due to the inability to activate IRF4 target genes involved in plasma cell differentiation.
(A) Mouse C75BL/6 splenic B cells were cultured with LPS + IL4; transduced with control retrovirus (MIG-ctrl), IRF4WT, or IRF4T95R; and analyzed for CD138 and B220 expression in transduced GFP+ cells. Top left: representative FACS profiles. The percentage of GFP+ cells is indicated. Left bottom: CD138 and B220 expression in gated GFP+ cells; the percentage of CD138+B220low cells is indicated. Right: summary of four independent experiments. Mean ± SD is shown. SSC-A, side scatter-A. (B) Spearman correlation coefficient between RNA-seq derived expression values of isolated mouse splenic B cells transduced with IRF4WT, IRF4T95R, or MIG-ctrl. (C) Top: number of differentially expressed (DE) genes in IRF4WT and IRF4T95R when compared with MIG-ctrl samples. Bottom: Genes differentially up-regulated by IRF4WT and IRF4T95R show limited overlap. (D) Comparison of genes differentially regulated by IRF4T95R with gene expression of selected lymphoid cell types. Genes associated with plasma cell differentiation are marked by a rectangle. (E to G) CH12 B cells were transduced with retrovirus expressing GFP alone, IRF4WT, or IRF4T95R and analyzed for the proportion of IgA+ cells in gated GFP+ cells 48 and 72 hours later. (E) Representative FACS profiles showing the virus-transduced GFP+ fraction (left) and IgA expression among the GFP+ cells (right). (F) Class switch to IgA at 48 and 72 hours after transduction of retrovirus expressing GFP alone, IRF4WT, or IRF4T95R. Mean ± SD of three independent experiments is shown. (G) Real-time PCR analysis of IRF4 and Aicda expression in sorted GFP+ CH12 cells after retrovirus transduction. The level of IRF4 and Aicda in CH12 cells expressing GFP alone was set to 1. (H) Generation of IRF4-deficient Ramos cells. Immunoblot for IRF4 protein expression in WT and IRF4-deficient Ramos clones 1-9 and 2-2 derived from 1# and 2# gRNA. (I and J) IRF4-deficient Ramos cells (clone 1-9) were transduced with retrovirus expressing GPF alone, IRF4WT, or IRF4T95R. The GFP+ cells were sorted 3 days later and analyzed for IgM-secreting cells by ELISPOT. (I) Left: Representative images of ELISPOT. Right: the number of IgM-secreting spots. Mean ± SD of triplicate wells is shown. (J) Sorted cells were further cultured for 5 days and analyzed by ELISA for IgM secreted into the culture supernatant. (K and L) Raji cells were transduced with retrovirus expressing GPF alone, IRF4WT, or IRF4T95R, and the GFP+ cells were sorted for RNA-seq. (K) Number of differentially expressed genes in Raji cells expressing IRF4WT or IRF4T95R as compared with Raji cells expressing GFP alone. (L) Expression heatmap depicting the differentially expressed genes shown in (K). Average transcripts per kilobase of exon model per million mapped reads (TPM) values of three independent samples standardized by z score are shown. Statistical significance was assessed by Tukey’s post hoc test (A, F, I, and J). *P < 0.05, **P < 0.01, and ****p < 0.0001.
To further define the B cell defect, we studied Ig class-switch recombination (CSR) in CH12 cells transduced with different IRF4 variants (WT and T95R) or GFP alone without further activation, and GFP+ cells were analyzed for the IgA+ population. IRF4T95R and GFP alone failed to induce IgA+ cells and failed to induce Aicda transcription (Fig. 5, E to G). To quantify plasma cell differentiation, we transduced the IRF4 variants into a human Burkitt’s lymphoma cell line, Raji, which endogenously expresses extremely low levels of IRF4. IRF4T95R failed to up-regulate genes encoding CD38, BLIMP1, and XBP1 (fig. S8, C to E). To assess the antibody secretion, we used the fact that Ramos cells (human Burkitt’s lymphoma cell line) spontaneously secrete low levels of IgM. After disrupting the endogenous IRF4 gene using CRISPR-Cas9–mediated genome editing (Fig. 5H), IRF4-deficient Ramos cells were transduced with the IRF4 variants, and IRF4T95R was unable to increase the frequency of antibody-secreting cells nor enhance IgM secretion (Fig. 5, I and J, and fig. S8, F and G). Co-expression of T95R with WT IRF4 in IRF4-deficient Ramos cells suppressed WT IRF4-induced plasma cell differentiation (fig. S8, H to J). In addition, ectopic expression of T95R, but not empty vector, suppressed the generation of CD138+ cells in human tonsil B cells activated with CD40L + IL21 (fig. S8K). These results revealed the antimorphic properties of T95R. To define the difference between IRF4WT and IRF4T95R at the transcriptome level, we performed RNA-seq on Raji cells expressing either version of IRF4. IRF4WT up-regulated 132 genes, including key regulators of B cell development such as PRDM1, XBP1, ERN1/IRE1, TNFRSF17, SLAMF7, and CD38 (Fig. 5, K to L). IRF4T95R up-regulated the expression of 38 genes, 19 of which are shared with IRF4WT. IRF4T95R did not hyperactivate the expression of anti-plasma cell TFs such as BCL6, BACH2, and ID3 (fig. S8L). Consistent with the predominant role of IRF4 as a transcriptional activator in B cells, few genes were down-regulated by either variant. When considered in aggregate, this series of experiments demonstrates that IRF4T95R is unable to induce CSR and plasma cell differentiation due to failure to up-regulate the expression of key target genes, including AICDA, PRDM1, and XBP1. However, IRF4T95R also demonstrated neomorphic functions by up-regulating a unique set of genes.
IRF4T95R has increased affinity for DNA, enhanced nuclear localization, and altered DNA binding specificity
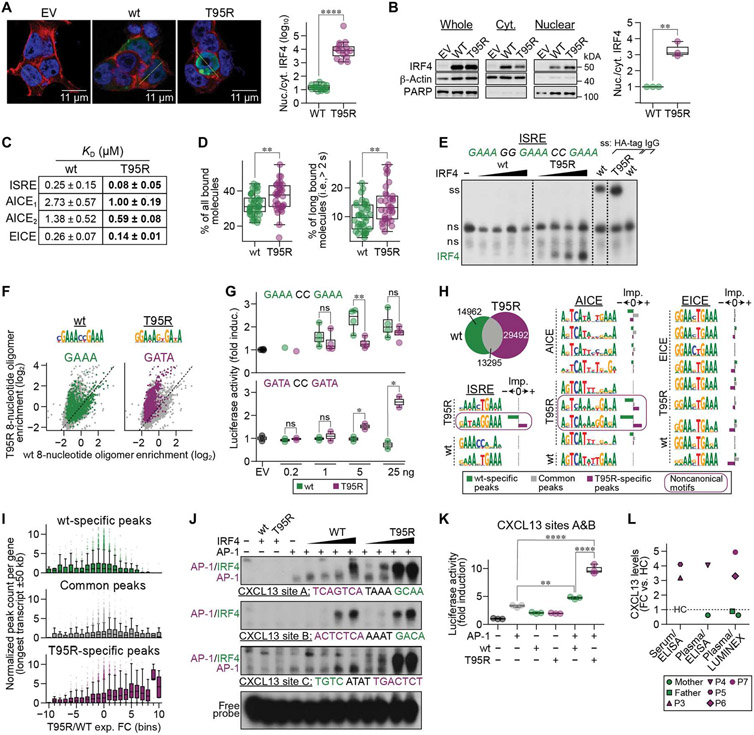
We next defined the intrinsic molecular mechanism of action of the IRF4T95R allele. Because of the additional positive charge introduced by the arginine, we hypothesized that IRF4T95R would have increased affinity for DNA (which is negatively charged) compared with IRF4WT. Moreover, protein affinity for DNA is associated with increased nuclear localization (19). Using both imaging and immunoblotting, we established that the subcellular localization of IRF4T95R was altered, with an increased nuclear-to-cytoplasmic ratio compared with IRF4WT (Fig. 6, A and B). Next, using surface plasmon resonance to quantify the interaction between the IRF4 and specific DNA sequences, we demonstrated that IRF4T95R has two- to threefold higher binding affinity [i.e., lower dissociation constant (KD)] than IRF4WT for the ISRE, AICE, and EICE motifs (Fig. 6C). This higher binding affinity of IRF4T95R was independently assessed by single-molecule fluorescence microscopy, showing that IRF4T95R makes both more and longer (i.e., specific) interactions with DNA than IRF4WT (Fig. 6D). Furthermore, electrophoretic mobility shift assays (EMSAs) biochemically confirmed the higher affinity of IRF4T95R for an ISRE DNA probe (Fig. 6E).
Fig. 6. IRF4T95R showed increased nuclear localization and affinity for DNA, altered specificity, and a different IRF4-binding landscape.
(A) 293T cells were transfected with an empty vector (EV) or a vector-expressing IRF4WT or IRF4T95R. Nuclei were stained with DAPI (blue), cytoplasm with phalloidin (red), and IRF4 with an anti-IRF4 antibody (green). Left: representative images. Right: a summary of randomly chosen cells. (B) Ratio of nuclear to cytoplasmic IRF4 in Raji cells transduced with retrovirus expressing IRF4WT or IRF4T95R. Left: representative immunoblot. Right: mean ± SD of three independent experiments. PARP, poly(adenosine diphosphate–ribose) polymerase. (C) IRF4T95R showed increased affinity for an ISRE, two AICEs, and an EICE site. (D) Fractions of all bound SiR-HaloTag- IRF4WT and IRF4T95R molecules (left) and molecules long bound for >2 s (right) as determined by single-molecule fluorescence microscopy with interlaced time-lapse illumination. (E) HEK293 cells were transfected with control plasmid (−), IRF4WT, or IRF4T95R. Nuclear extracts were analyzed by EMSA using 3xGAAA ISRE. Supershifts (ss) of WT and T95R extracts using HA-tag antibody or IgG control are shown at the far right. Note that IRF4T95R binds more strongly to ISRE compared with IRF4WT. Dashed lines indicate cuts of the scan for presentation. (F) Top: IRF4WT (left) and IRF4T95R (right) motifs found in the HT-SELEX data. Bottom: 8-nucleotide oligomer containing GAAA (left) or GATA (right) enriched in IRF4WT (x axis) or IRF4T95R (y axis). (G) 293T cells were transfected with a TK-cypridina luciferase vector (an internal control) and either a canonical (ISRE)1–driven luciferase vector (top) or a noncanonical (ISRE)1–driven luciferase vector (bottom), together with a pFLAG-CMV empty vector (400 ng) or increasing amounts of plasmids encoding IRF4WT or IRF4T95R. The luciferase activity was compared with that induced by the empty vector, which was set to 1. Mean ± SD of two to four independent experiments is shown. (H) ChIP-seq analysis of immortalized B cells from P3 compared with a HC. Top left: overlay of IRF4 ChiP-seq peaks in EBV-B cells of P3 and HC. From left to right: ISRE, AICE, and EICE motifs found in IRF4T95R, IRF4WT, AICE, or EICE ChIP-seq data (indicated at the left of the motifs). The importance of each motif toward the IRF4T95R-specific (purple), IRF4WT-specific (green), or common (gray) component of the ChIP-seq data is shown to the right of each motif. Noncanonical motifs are surrounded by a purple line. (I) Normalized IRF4WT-specific (green, top), common (gray, middle), and IRF4T95R-specific (purple, bottom) ChIP-seq peak counts (y axis) for different groups of differentially expressed genes. (J) HEK293 cells were transfected with AP-1 (JUNB and BATF) with or without IRF4WT or IRF4T95R, as indicated. Nuclear extracts were analyzed for binding to various CXCL13 sites, as indicated. Note that IRF4T95R shows strongly increased (CXCL13-A) or exclusive (CXCL13-C) binding compared with IRF4WT. (K) HEK293 cells were transfected with CXCL13 reporter construct encompassing CXCL13 sites A and B together with AP-1 (JUNB and BATF) and IRF4 variants, as indicated. Luciferase activity is shown as fold activation compared with control transfected cells (far left), which is set as 1. Mean ± SD of three independent experiments is shown. (L) FC of CXCL13 levels in serum or plasma from P3 to P7 compared with HC. Statistical significance was determined by one-tailed Welch’s t test (A, B, D, and G) and by Tukey’s post hoc test (K). *P < 0.05, **P < 0.01, and ****p < 0.0001.
Alignment of the nine human IRF protein sequences revealed that the arginine at position 95 of IRF4T95R is analogous to the arginine at position 78 of IRF3 (fig. S9A). This arginine allows IRF3 to bind to nonconsensus DNA sites (20). Hence, we hypothesized that IRF4T95R would also be able to bind to nonconsensus DNA sites. High-throughput systematic evolution of ligands by exponential enrichment (HT-SELEX) analysis revealed that, in addition to binding to the consensus IRF GAAA sequence, IRF4T95R had gained the ability to bind to GATA-containing sequences (Fig. 6F). This neomorphic ability of IRF4T95R to bind to GATA sequences was verified by EMSA, which also demonstrated that IRF4WT is unable to bind to this sequence (fig. S9B). Together, IRF4T95R showed an increased nuclear localization paired with increased DNA binding affinity and the ability to target additional DNA motifs that were not recognized by IRF4WT, further supporting the hypermorphic and neomorphic functions associated with IRF4T95R.
Although IRF4T95R could bind to both GAAA- and GATA-containing ISRE sites, it remained unclear whether it could actually activate target gene transcription. To test this possibility, we performed luciferase reporter assays to assess the ability of IRF4WT and IRF4T95R to activate transcription from reporter constructs containing a canonical (GAAACCGAAA) or a noncanonical ISRE (GATACCGATA). Although IRF4WT activated the canonical ISRE construct in a dose-dependent fashion, IRF4T95R exhibited hypomorphic activity (Fig. 6G, top). However, confirming IRF4T95R neomorphic activity, IRF4T95R activated the GATA-containing construct in a dose-dependent fashion, whereas IRF4WT could not activate this construct at all (Fig. 6G, bottom).
T95R changes both the genome-wide binding landscape of IRF4 and the transcriptome
To evaluate the genome-wide landscape of IRF4 binding in patient cells, we performed IRF4 chromatin immunoprecipitation followed by sequencing (ChIP-seq) of immortalized EBV-B cells from an IRF4T95R patient and an IRF4WT healthy control (data file S2). Overall, IRF4T95R B cells had more IRF4 ChIP-seq peaks than IRF4WT B cells (Fig. 6H). Emphasizing the neomorphic activity of IRF4T95R, more than 35% of the peaks (versus <10% in IRF4WT) corresponded to “non-ChIPable” IRF4 regions [i.e., they are not reported in the ReMap database (21), which aggregates IRF4 ChIP-seq data from B cells, T cells, plasmablasts, and various cell lines], and about 33% do not overlap any of the >1 million candidate cisregulatory elements from ENCODE (22) (versus ~7% in IRF4WT). Applying a new deep learning tool, ExplaiNN (explainable neural networks) (23), we separately identified motifs de novo in four different datasets, including the patient and healthy control ChIP-seq datasets and two custom datasets describing the binding of IRF4 to either AICE or EICE sites in GM12878 cells. Next, we used these motifs to initialize a “surrogate” ExplaiNN model in a process known as transfer learning with which to evaluate their importance toward the IRF4T95R-specific, IRF4WT-specific, or common component of the ChIP-seq data (fig. S9C and full data in data file S3). In the patient dataset, ExplaiNN identified a noncanonical, GATA-containing ISRE motif, matching the results of the HT-SELEX analysis, and various noncanonical AICE motifs that were important for IRF4T95R-specific peaks but detrimental for IRF4WT-specific peaks (Fig. 6H). In contrast, identified EICE motifs in the different datasets were regarded as detrimental for IRF4T95R-specific peaks compared with IRF4WT-specific and common peaks (Fig. 6H). PU.1 cofactor-independent binding of IRF4T95R was observed at EICE; however, increasing amounts of IRF4T95R outcompeted the formation of PU.1-IRF4T95R composite complexes at this site (fig. S9D), providing a possible explanation for the irrelevance of EICE motifs in the patient-derived ChIP-seq peaks. Furthermore, increased co-factor-independent IRF4T95R binding to EICE depended on the presence of a C in position +1 in relation to the GAAA motif (fig. S9E), suggesting that nucleotides adjacent to the core-binding motif influence IRF4T95R binding capacity. Together, these data confirmed the neomorphic DNA binding capacity of IRF4T95R to noncanonical ISRE and noncanonical AICE sites in primary patient cells.
Next, through matching of RNA-seq and ChIP-seq data on the same cells, we found an association between IRF4 binding and gene expression levels (Fig. 6I). Specifically, genes that were highly expressed in IRF4T95R patient cells compared with IRF4WT cells had more IRF4T95R ChIP-seq peaks around them (longest transcript ±50 kb) (Fig. 6Iand data file S2). To validate the functional impact of this neomorphic binding activity of IRF4T95R, we focused on CXCL13. CXCL13 is a key chemokine involved in GC activity and lymph node architecture (24), and CXCL13 was the most differentially expressed gene in patient IRF4T95R B cells when compared with healthy controls (log2 expression fold change = 11.98; data file S2). The ±50-kb region around the longest CXCL13 transcript (hg38:chr4:77,511,752 to 77,611,834) harbored 15 IRF4T95R peaks, of which two overlapped with the only two IRF4WT peaks in the region (fig. S9F). Two of the 15 IRF4T95R peaks encompassed noncanonical AICE sites (designated as CXCL13 sites A, B, and C), none of which contained the canonical GAAA half-ISRE motif. EMSA analyses confirmed strongly increased (CXCL13 site A) or exclusive (CXCL13 site C) neomorph binding of IRF4T95R compared with IRF4WT in the presence of the AP-1 factors JUNB and basic leucine zipper transcriptional factor ATF-like (BATF) (Fig. 6J and fig. S9G). However, at CXCL13 site B, which is located within the same peak and only 81 bp away from site A, both IRF4T95R and IRF4WT showed similar binding patterns. To investigate the functional impact of this binding pattern, we analyzed the activity of a luciferase reporter construct containing the two AICE sites, CXCL13 site A and CXCL13 site B (Fig. 6K). Whereas IRF4WT only slightly increased luciferase activity above the AP-1–induced activity, IRF4T95R induced robust luciferase activity in the presence of AP-1. Last, to analyze whether this IRF4T95R-mediated transcriptional increase is mirrored by biochemical changes in vivo, we measured CXCL13 serum or plasma levels of five IRF4T95R patients and found that they had consistently 3- to 4.5-fold increased CXCL13 levels compared with healthy controls (Fig. 6L). In combination, these data confirm the functional impact of the neomorphic IRF4T95R binding activity.
DISCUSSION
Foundational to our understanding of genetics are “Muller’s morphs,” where Muller suggested that a mutant allele can be classified into one of five types: amorph, hypomorph, hypermorph, neomorph, and antimorph (25). Here we have defined a previously undescribed mechanism of human disease caused by the “multi-morphic” activity of IRF4T95R. The T95R amino acid change results in an unanticipated combination of hypermorphic, hypomorphic, and neomorphic functions in the TF, causing the clinical and immunological phenotype we document in the seven patients with CID. IRF4T95R behaves as a gain-of-function hypermorph by binding to DNA with higher affinity than IRF4WT. Despite this increased affinity for DNA, the transcriptional activity on IRF4 canonical genes is reduced, showcasing the hypomorphic activity of IRF4T95R. Simultaneously, IRF4T95R functions as a neomorph by binding to noncanonical DNA sites to alter the gene expression profile, including the transcription of genes exclusively induced by IRF4T95R but not by IRF4WT. In combination, this multimorphic activity of the IRF4T95R variant leads to a transcriptional space, which is detrimental to activation and differentiation of immune cells.
Similar neomorphic functions have been reported in other TFs. In most cases, these neomorphs and the ensuing altered expression of genes predisposes to malignancies (26-28). Furthermore, neomorphic germline mutations cause osteogenesis imperfecta in SP7 (29), anemia in EKLF (30), and arteriopathy in NOTCH3 (31), to mention a few. However, we are not aware of human diseases caused by the multimorphic combination of hypermorphic, hypomorphic, and neomorphic activity as displayed by IRF4T95R. Overall, our findings highlight a distinct molecular mechanism for human disease and expand the spectrum of diseases originated from mutations in IRF4.
The heterozygous IRF4T95R variant found in multiple unrelated families caused a fully penetrant, severe very early-onset immunodeficiency characterized by greatly enhanced susceptibility to opportunistic pathogens such as P. jirovecii and weakly pathogenic mycobacteria. In contrast, IRF4 haploinsufficiency caused by the heterozygous loss-of-function variant IRF4R98W found in a single family was associated with age-dependent, incomplete penetrance of Whipple’s disease, an inflammatory intestinal disease caused by Tropheryma whipplei (32). Early and severe disease onset was also reported in a single IRF4-deficient patient carrying a homozygous splicing mutation in IRF4 due to uniparental isodisomy (33). Given the very complex genetics of this isolated case of IRF4 deficiency, the possibility of other contributing factors cannot be excluded. Nevertheless, some immunological features are shared between the heterozygous IRF4T95R patients we describe here, the IRF4-deficient patient (33), and Irf4 knockout mice (3, 34), including low percentages of memory B lymphocytes in peripheral blood, very low levels of plasma cells in the bone marrow or lymph nodes, agammaglobulinemia, polyclonal T cell repertoire, and low percentages of TH17 and TFH cells.
Our analysis of heterozygous p.T95R Irf4 knock-in mice recapitulates the human phenotype and further establishes the causal role of the IRF4T95R mutation. The reduced production of antibodies after immunizations and infections and the absence of plasma cells after in vitro differentiation point to a failure of IRF4T95R to support terminal B cell differentiation. However, our finding that Irf4T95R/+ mice have reduced formation of antigen-specific GC B cells, despite an overall increase in the number of GC B cells observed in naïve Irf4T95R/+ mice, suggests a broader dysregulation of the GC reaction and a very different mechanism for the lack of high-affinity antibodies than a simple loss of IRF4. Together, our data suggest that altered subcellular localization, higher DNA binding affinity, and noncanonical binding of IRF4T95R perturb the dynamic spatial and temporal expression of IRF4, resulting in dysregulated GC reactions, impaired Ig CSR, disturbed T cell differentiation, and decreased plasma cell differentiation.
In conclusion, we describe a human immune disorder caused by the heterozygous germline IRF4T95R mutation. The simultaneous multimorphic combination of dominant loss, gain, and new functions for IRF4T95R represents a new mechanism for human disease. We suggest that this new human disease be named “multimorphic IRF4 CID” (or MICI). Because IRF4 is primarily expressed by cells of the immune system, IRF4T95R manifests disease as a CID. However, we anticipate that variants with multimorphic activity may be more widespread in health and disease, particularly variants in TFs that alter the genome-wide binding and transcriptional landscapes.
MATERIALS AND METHODS
Study design
The objective of this study was to elucidate the pathophysiology of a combined immunodeficiency caused by a recurrent, de novo missense heterozygous IRF4 variant. We performed extensive phenotyping of the patients’ peripheral blood cells by CyTOF, scRNA-seq, and conventional flow cytometry to reveal the immune cell abnormalities associated with the disease. Functional defects of B cells, T cells, and myeloid cells were then analyzed by inducing their activation and differentiation under in vitro culture conditions. We further generated knock-in mice with the identical T95R substitution and confirmed that the IRF4T95R heterozygous mice recapitulated the immunodeficient phenotypes observed in these patients. We uncovered the altered DNA binding specificity and transcriptional activity of IRF4T95R by multiple approaches, including surface plasmon resonance, single-molecule fluorescence microscopy, EMSA, HT-SELEX, luciferase assay, ChIP-seq, and a deep learning tool (ExplaiNN).
Patients and healthy controls
All study participants and/or their parents/guardians provided written informed consent. Research study protocols were approved by local institutions, specifically the University of British Columbia Clinical Research Ethics Board (H15-00641 and H18-02853); ethics committee of the Children’s Hospital of Fudan University (registration no. NCT03383380); Comité de Protection des Personnes Ile de France II, Paris, France (reference: 2015-01-05; 2015-01-05 MS2); the Vanderbilt University Medical Center Human Research Protection Program (Institutional Review Board no. 200412); the French Advisory Committee on Data Processing in Medical Research (Comité Consultatif sur le Traitement de l’lnformation en matière de Recherche dans le domaine de la Santé, Paris, France; reference: 15.297bis); the University of Ulm ethics board, Germany (application number 156/11); and the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH).
Mice (Shanghai)
Irf4T95R mutant mice and conventional Irf4−/− mice were generated by CRISPR-Cas9-mediated genome engineering. For the Irf4T95R mutant allele, a guide RNA (gRNA) that targets exon 3 of the Irf4 gene and single-stranded donor oligonucleotides were synthesized at Integrated DNA Technologies. The gRNA, Cas9, and donor oligo were coinjected into C57BL/6 fertilized eggs (Cyagen, Suzhou, China). The sequences of the gRNA and donor oligo are as follows: gRNA, 5′-CGTAATCTTGTCTTCCAAGT-3′ and donor oligo, 5′-GGCTTGGGCATTGTTTAAAGGCAAGTTCCGAGAAGGGATCGACAAGCCAGATCCTCCTACGTGGAAGAGAAGATTACGATGTGCTCTGAACAAGAGCAATGACTTTGAGGAATTGGTCGAGAGGAGCCAG-3′. For conventional Irf4 knockout, double gRNAs were designed to delete exons 3 to 6 of the Irf4 gene. The sequences of gRNAs are as follows: gRNA1, 5′-AGGGACTCACACGGGGTCGG-3′ and gRNA2, 5′-TGTCTGCTTCCACGGAGTGT-3′. The gRNAs and Cas9 were coinjected into C57BL/6 fertilized eggs (Cyagen, Suzhou, China).
Male and female mice at 8 to 12 weeks of age were used for the functional and biochemical analysis. All mouse strains were bred and maintained in the specific pathogen–free animal facility of Fudan University. All animal studies were conducted in accordance with the Animal Experiment Committee of Fudan University and used the committee’s approved protocols.
Mice (ANU)
Irf4T9SR and Irf4−/− mice were generated by CRISPR-Cas9–mediated genome engineering in an in-house facility. For generation of the T95R point mutation, gRNA1 (5′-CGTAATCTTGTCTTCCAAGT-3′) and a single-stranded donor Oligo (5′-TCTGGAACAATCCTGTACACCTTGTATGGGTCAGAGATATCCAGCTGGCTCCTCTCGACCAATTCCTCAAAGTCATTGCTCTTGTTCAGAGCACATCGTAATCTTCTCTTCCAAGTAGGAGGATCTGGCTTGTCGATCCCTTCTCGGAACTTGCCTTTAAACAATGCCCAAGCCTAAGAACGATGAGA-3′) from Integrated DNA Technologies were coinjected into fertilized C57BL6/N zygotes [Cas9 (50 ng/μl),gRNA (10 ng/μl), and ss oligo (100 ng/μl)] in a form of a ribonucleoprotein complex. For generation of Irf4−/−, mice gRNA1 (5′- CGTAATCTTGTCTTCCAAGT-3′) and gRNA2 (5′-GAACAAGAGCAATGACTTTG-3′) were coinjected into fertilized C57BL/6N zygotes [Cas9 (50 ng/μl) and sgRNA (10 ng/μl)]. Irf4−/− mice contain a 47–base pair (bp) deletion leading to a frame shift from AA92 of the IRF4 protein and a premature stop codon. Lack of protein expression was confirmed by Western blot (fig. S6A).
C57BL/6 mice, Irf4T95R/+, and Irf4+/− were maintained on a C57BL/6N background and bred in pathogen-free conditions at the Australia National University (ANU, ACT, Australia) Phenomics Facility. Unless stated otherwise, all mice used were aged between 8 and 12 weeks. All animal experiments were approved by the ANU Animal Experimentation Ethics Committee on protocols A2014/058, A2017/54, and A2020/45 according to the National Health and Medical Research Council Australian code of practice.
Cells
Human embryonic kidney (HEK) 293T, Ampho, Raji, and Ramos cells were obtained from the American Type Culture Collection. CH12 cells were provided by T. Honjo (Kyoto University).
Genetic analysis
Genomic DNA was extracted from the whole blood or PBMCs and fibroblasts of the patients and their parents. Next-generation sequencing of the genomic DNA was performed using an Illumina sequencing platform using either whole exome or a targeted panel approach. Bioinformatics analysis for detection of rare sequence variants was performed as described previously (35, 36).
Mutations in IRF4 were confirmed using Sanger sequencing. Primers used for sequencing are listed in table S13.
Functional predictions of the IRF4 variant
Predictions of the functional relevance of the detected IRF4 variant were performed using the dbNSFPv4 database (https://sites.google.com/site/jpopgen/dbNSFP) (37).
Measurement of Ig plasma levels
Plasma IgG, IgA, and IgM levels were determined by an automated clinical chemistry analyzer (Erba, model: XL-200) as previously described (36) or by immunoturbidimetric assays (Roche Diagnostics, Switzerland). Serum IgE was measured by UniCAP (Pharmacia).
Cell isolation and culture
Human CD4+ and CD8+ naïve T cells (defined as CD45RA+ CCR7+ or CD45RA+) were isolated (>98% purity) with a FACSAria II cell sorter (BD Biosciences) or using the MojoSortTM Human CD4 Naïve T Cell Isolation Kit (BioLegend, USA) according to the user manual. Human naïve B cells were isolated from PBMCs using a human naïve B cell enrichment kit (STEMCELL Technologies) or the MojoSort Human Naïve B Cell Isolation Kit (BioLegend) together with biotinylated anti-human IgG beads to remove IgG+ cells or human pan B cell isolation kit (BioLegend) [memory B cells were removed by a second isolation step using biotinylated anti-human CD27 and F(ab′)2 anti-human IgG and IgA antibodies binding to streptavidin-coupled paramagnetic beads and a Mojo magnet as described (38-41)]. Mouse bone marrow cells were flushed from femurs and tibias with cold fluorescence-activated cell sorting (FACS) buffer [phosphate-buffered saline (PBS) supplemented with 2% FBS], and spleen and Peyer’s patch cells were obtained by gently teasing these tissues onto a 70-μm cell strainer. Spleen B cells were purified using a negative sorting kit (BD Biosciences) or a magnetic-activated cell sorting B cell isolation kit (Miltenyi Biotec). Erythrocytes were removed by using ACK (ammonium-chloride-potassium) buffer.
Purified human naïve B cells, human naïve T cells, human PBMCs, mouse purified spleen B cells, Raji cells, Ramos cells, and CH12 cells were cultured in RPMI 1640 (Gibco) containing 5 × 10−5 M2-mercaptoethanol (Sigma-Aldrich), penicillin (100 U/ ml), and streptomycin (100 μg/ml) (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco). Stimuli were freshly added as described in the figure and figure legends. 293T and Ampho cells were cultured in Dulbecco’s modified Eagle medium (Gibco) supplemented with 10% FBS. All cells were cultured at 37°C in humidified air and 5% CO2.
For in vitro plasma cell differentiation, enriched human naïve B cells were activated with F(ab′)2 anti-human (2 μg/ml) plus 0.6 μM CpGODN 2006 and IL2 (10 ng/ml; CMIL2) or CD40L (200 ng/ml) and IL21 (50 ng/ml). Mouse B cells were activated with 0.1 volume of CD40L plus IL4 (20 ng/ml) and IL21 (20 ng/ml) or LPS (20 μg/ml).
For the induction of the differentiation into TFH and Tfr cells, purified naïve human CD4+ T cells were incubated in AIM-V medium (Gibco, USA) supplemented with IL12 (5 ng/ml; Pepro-Tech, Germany), Activin A (100 ng/ml; PeproTech, Germany) and one bead per cell Dynabeads Human T-Activator CD3/CD28 (Gibco, USA) for the specified times (42).
Establishment of EBV-B cell lines
EBV-B immortalization was performed as previously reported (43).
Cytometry by time of flight
The Maxpar direct immune profiling assay (Fluidigm, #201325) in combination with Maxpar direct T cell expansion panel 2 (Fluidigm, #201406) was used for the high-dimensional immune profiling of PBMCs. For each sample, cells were thawed, washed once, and checked for viability (>80%). After a 10-min incubation with an FC blocker (TruStain FCX, BioLegend) in MaxPar staining buffer, cells were directly transferred into the antibody-containing tube. Cells were then incubated for 10 min in 1.6% formaldehyde solution, washed once, transferred into an intercalator ID solution, and incubated overnight at 4°C. Immediately before acquisition, the cells were washed, resuspended in Maxpar cell acquisition solution (1 million cells/ml), and mixed with 10% (v/v) EQ calibration beads. An average of 500,000 events were acquired per sample on a Helios mass cytometer. The acquisition data were analyzed with CyTOF software (version 6.7.1014, Fluidigm), enabling Maxpar-Pathsetter automated single-cell analysis at the Cytometry Facility in Pitié-Salpetriere Hospital (Paris, France).
For multidimensional analysis, Flow Cytometry Standard (FCS) files were pregated in Flowjo 10.7.2 (Becton, Dickinson, and Company, Ashland, OR) following standard procedures (44). The pregated data were then imported into RStudio and analyzed using the package CATALYST (45). Unsupervised clustering and cluster characterization were performed with FlowSOM (46) and marker enrichment modeling (MEM) (47). Additional gating was performed in Flowjo. We used ggplot2 and Prism 9 (GraphPad Software, San Diego, CA) for data representation.
Single-cell RNA sequencing
To compare gene expression in single B cells, we sorted CD19+ cells from PBMCs using MoFlo XDP (Beckman). Either CD19+ B cells or PBMCs were processed using the 10x Genomic Chromium Single Cell Platform at a concentration of 700 to 1000 cells/μl as described in the manufacturer’s protocol. About 8000 to 10,000 cells were loaded onto 30 library chips following the Single Cell 30 Reagent Kits (v2) User Guide. The cells were then partitioned into gel beads in emulsion in the GemCode instrument, where cell lysis and barcoded reverse transcription of RNA occurred, followed by amplification, enzymatic fragmentation, 50 adaptor, and sample index attachment. The libraries were constructed using Chromium Single Cell 30 Reagent Kits (v2), Single Cell 30 Library and Gel Bead Kit v2 (PN-120237), Single Cell 30 Chip Kit v2 (PN-120236), and i7 Multiplex Kit (PN-120262) (10x Genomics). The generated scRNA-seq libraries were sequenced using an Illumina HiSeq 4000 platform as 150-bp paired-end reads at one full lane per sample.
Shanghai NovelBio Co. Ltd. performed scRNA-seq data analysis using the NovelBrain Cloud Analysis Platform. scRNA-seq reads were aligned to the Genome Reference Consortium Human Build 38 (GRCh38), normalized for batch effects, and filtered for cell events using the Cell Ranger software (v3.0.0). Cells expressing fewer than 200 genes and cells with a mitochondrial unique molecular identifiers (UMI) rate higher than 20% were excluded. Mitochondrial genes were removed from the expression matrix but used for cell expression regression to avoid the cell status effect for clustering analysis and marker analysis of each cluster. The Seurat package (version: 2.3.4) was used for normalization based on the expression matrix according to the UMI counts of each sample and percentage of mitochondria rate to obtain the scaled data. Using the graph-based cluster method, we acquired the unsupervised cell cluster results on the basis of the principal components analysis top 20 principal. To identify differentially expressed genes between the control and disease groups of the same cell type, we used the Seurat package FindMarkers function using the Wilcox rank-sum test algorithm under the following criteria: (i) log fold change (logFC) > 0.25, (ii) P value < 0.05, and (iii) min.pct > 0.1.
Immunohistochemistry of lymph node tissue
Immunostaining of formalin-fixed and paraffin-embedded tissue sections (2 μm) were carried out by the avidin-biotin-complex method (48). The antibodies used are listed in table S12.
Mouse immunizations
For humoral immune responses, mice were immunized intraperitoneally with 25 μg of NP-CGG (Biosearch Technologies) in 4.5% alum in 200 μl of PBS. NP-specific IgM and IgG1 antibodies were measured by enzyme-linked immunosorbent assay (ELISA) as described (49). Antibody responses to CGG and formalin-fixed B. pertussis in mice were determined 14 days after immunization as described previously (50).
GC analysis in mice
For induction of GC reactions, each mouse was intravenously injected with 200 μl of the SRBC suspension containing 2 × 108 SRBCs, and spleens were analyzed at day 10. Alternatively, Plasmodium berghei parasites engineered to express Plasmodium falciparum CSP (PfCSP) in place of the endogenous P. berghei CSP molecule (Pb-PfSPZ) (51) were used. Parasites were maintained by serial passage through Anopheles stephensi mosquitoes. Mice were immunized intravenously with 5 × 104 irradiated (15 kRad) Pb-PfSPZ dissected by hand from the salivary glands of A. stephensi mosquitoes as described previously.
For the detection of PfCSP-specific cells, a nine-times repeat of asparagine-alanine-asparagine-proline (NANP)9 peptide was sourced from Biomatik (Ontario, Canada) and was biotinylated with the Sulfo-NHS-LC Biotinylation Kit (Thermo Fisher Scientific) at a ratio of 1:1 according to the manufacturer’s instructions. Biotinylated antigens were incubated with premium-grade streptavidin-phycoerythrin and streptavidin-allophycocyanine (Molecular Probes) at a molar ratio of 4:1 and added four times with 15-min incubation at room temperature.
Quantitative PCR
Cells were lysed in RNAiso Plus (TaKaRa) and stored at −80°C. Total RNA was extracted following the manufacturer’s instructions. cDNA was synthesized using the First Strand cDNA Synthesis Kit (Yeasen). Quantitative polymerase chain reaction (PCR) was performed using a Light Cycler 480 Instrument II (Roche) with TB Green Premix Ex Taq (TaKaRa). Primers used for quantitative PCR are listed in table S13.
Establishment of IRF4-deficient Ramos cells
Two gRNAs were designed to target genomic DNA within the second and third exons of the IRF4 gene by online software (www.crispr-cas.org/), which predicted high-specificity and protospacer adjacent motif target sites in the human exome. Construction of lentiCRISPR vector and collection of lentivirus have been described (52). Ramos cells were cultured in the presence of viral supernatant and selected for the correctly targeted clones as described (52).
ELISPOT assay
This assay was performed as described previously (49). Briefly, multiscreen high-throughput screening plates (Millipore) were coated with goat anti-human Ig (50 μg/ml; SouthernBiotech) or rabbit anti-mouse Ig (SouthernBiotech). Serially diluted cells were added to individual wells in triplicate and then incubated at 37°C for 2 hours in a CO2 incubator. The plates were further incubated with biotin–anti-human IgM or IgG (SouthernBiotech) followed by alkaline phosphatase–conjugated streptavidin (SouthernBiotech) or directly with AP-conjugated anti-mouse IgM or IgG1 (Southern-Biotech). Spots were revealed by 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium reagent (MOSS Inc.), and colonies were counted using an enzyme-linked immune absorbent spot (ELISPOT) reader (Autoimmun Diagnostika GmbH).
RNA-seq of splenic B cells
RNA-seq data from mouse splenic B cells were processed using PiGx RNA-seq pipeline (53). In short, the data were mapped onto the GRCm38/mm10 version of the mouse transcriptome [downloaded from the ENSEMBL database (54)] using SALMON (55). The quantified data were processed using tximport (56), and the differential expression analysis was done using DESeq2 (57). Genes with fewer than five reads in all biological replicates of one condition were filtered out before the analysis. Two groups of differentially expressed genes were defined—a relaxed set containing genes with an absolute log2 expression FC of 0.5 and a stringent set containing games with an absolute log2 expression FC of 1. The FC was deemed significant if the adjusted P value was less than 0.05 (Benjamini-Hochberg corrected).
Splenic B cell per sample heatmap was constructed by calculating the pairwise Pearson correlation coefficient between samples. The expression values were normalized using DESeq2. The heatmap was visualized using the ComplexHeatmap package (58). Human and mouse genes were mapped through the orthologous assignment using the ENSEMBL database. Monocyte, B cell, and plasma cell expression profiles were extracted from the ARCHS4 database (59).
RNA-seq of Raji cells
Total RNA was extracted using RNAiso Plus (TaKaRa) from Raji cells transduced with retrovirus-expressing WT or mutant IRF4. RNA-seq was performed by BGI (Beijing Genomic Institute, Shen- Zhen, China) using the BGISEQ-500 platform, paired-end 100 bases read lengths. The sequencing data were filtered with SOAP-nuke (v1.5.2). The clean reads were mapped to the reference genome using HISAT2 (v2.0.4). After that, Ericscript (v0.5.5) and rMATS (V3.2.5) were used to identify genes and differentially spliced genes, respectively. Bowtie2 (v2.2.5) was applied to align the clean reads to the gene set, a database for this organism built by BGI with known and novel coding transcripts included, and then the expression level of each gene was calculated by RSEM (v1.2.12). Heatmaps were drawn by pheatmap (v1.0.8) according to the gene expression levels in different samples. Essentially, differential expression analysis was performed using the DESeq2 (v1.4.5) under the following criteria: (i) logFC > 1, (ii) q value < 0.001, and (3) read counts > 10 and fragments per kilobase of exon model per million mapped fragments (FPKM) > 0.5.
Immunofluorescence
Transiently transfected HEK293T cells were fixed by incubation with 4% paraformaldehyde for 15 min on coverslips and permeabilized in a 10-min incubation with 1× PBS–5% BSA/0.1% Triton X-100. Cells were washed with filtered PBS, blocked by a 40-min incubation with PBS–5% BSA, stained for IRF4 (Cell Signaling, #4948) for 1 hour, washed, and incubated with a secondary antibody (anti-rabbit fluorescein isothiocyanate, Jackson ImmunoResearch, #50784). 4′,6-diamidino-2-phenylindole (DAPI; Chemometech, #910-3018) and phalloidin (Cell Signaling, #8940) were added for 40 min. After several washes, fluorescence was detected with a confocal Leica SP8 microscope. Images were analyzed with Fiji software.
HT-SELEX
HT-SELEX with the randomized 40 bp–based synthetic ligand was performed as described in (60). To compute 8-nucleotide oligomer (i.e., DNA sequences with a length of 8) enrichments, for each dataset, we obtained the number of occurrences of each 8-nucleotide oligomer in the first and last SELEX cycles was obtained using Jellyfish (version 2.2.10) (61). Then, the enrichment of each 8-nucleotide oligomer was computed as the logarithm to the base 2, resulting from the division between the number of occurrences of that 8-nucleotide oligomer in the last and first SELEX cycles. Motifs were obtained using ExplaiNN (23) (see the “Deep learning models” section in the Supplementary Materials and Methods).
ChIP-seq
For ChIP-seq analysis, 10 × 106 EBV-B cells from P3 and a healthy control C1 were fixed with freshly prepared 1% formaldehyde (Sigma-Aldrich, #F-8775) for 15 min, quenched with 0.125 M glycine (Sigma-Aldrich, #G-7403) for 5 min, and washed twice with 1× PBS containing 0.5% IGEPAL CA-630 (Sigma-Aldrich, #I-8896) and 1 mM phenylmethanesulfonyl fluoride (in the second wash only). Cell pellets were snap-frozen and subjected to ChIP-seq. Chromatin extraction, immunoprecipitation with an anti-IRF4 antibody (Cell Signaling, #4948), library preparation, next-generation sequencing, and a model-based analysis of the ChIP-seq data (47) were performed by Active Motif.
Matched ChIP-/RNA-seq analysis
Genes were grouped into 21 different bins, ranging from −10 to +10 according to their log2 expression FC in patient (i.e., IRF4T95R) versus healthy control cells (i.e., IRF4WT; data file S2). For example, genes whose log2 expression FC was smaller than −9.5 were assigned to the “−10” bin, between −9.5 and − 8.5 to the “−9” bin, between −8.5 and − 7.5 to “−8” bin, etc. For each gene, the number of IRF4T95R-specific, IRF4WT-specific, and common ChIP-seq peaks were obtained using BEDTools intersect on the genomic window encompassing the longest transcript of that gene, according to RefSeq definitions (62), ±50 kb. Peak counts were further normalized by applying a normalization factor, which, for each gene, was obtained by dividing the median genomic window size across all genes (i.e., 127,269 bp) by the genomic window size of that gene.
Electrophoretic mobility shift assay
Preparation of whole cell and nuclear extracts as well as Western blotting was performed as previously described (63, 64). Custom single-stranded IRDye 700–labeled oligonucleotides were ordered from Integrated DNA Technologies, and double-stranded oligonucleotides were generated by annealing in Tris-EDTA (TE) buffer (pH 7.9) supplemented with 33.3 mM NaCl and 0.67 mM MgCl2. The annealing conditions were 95°C for 3 min followed by cooling overnight at room temperature. Supershift assays were performed with 15 μg of nuclear protein lysate incubated on ice for 30 min with either IRF4 (#4964S, Cell Signaling) or IgG (611-145-002, Rockland) or hemagglutinin (HA)–tag (C29F4, Cell Signaling) antibodies and then incubated at room temperature for 20 min with probes shown in table S13. Protein-oligonucleotide-antibody mixtures were then subjected to electrophoresis in 5% acrylamide/Bisacrylamide 19:1 gels in 1% Tris-borate EDTA migration buffer for 60 min at 70 V. A LI-COR Odyssey infrared imager (LI-COR Bioscience) was used for imaging.
Luciferase reporter assay
For canonical and noncanonical ISRE reporter assay, HEK293T cells in a 24-well plate were transiently transfected with 500 ng of a 1xISRE reporter plasmid (which contains one ISRE sequence) or pGL3 basic backbone plasmid, thymidine kinase (TK)-cypridina vector (an internal control, 50 ng/well), and increasing amounts of pFLAG-CMV-5a vector expressing WT or mutant IRF4 using the Hieff Trans Liposomal Transfection Reagent (Yeasen). Cells were harvested 24 hours after the transfection, and the luciferase activity was measured with the Dual Luciferase Reporter Gene Assay Kit (Beyotime) according to the manufacturer’s protocol. Each transfection was performed in duplicate, and reporter activity is expressed as fold induction relative to cells transfected with the empty vector. At least three independent experiments were performed for each reporter plasmid. For the CXCL13 reporter assay, a gBlock DNA fragment was cloned into the pGL3-Basic plasmid (Promega, USA) using the Sma I restriction site. The sequences of the DNA fragment were listed in table S13. The plasmid was checked by Sanger sequencing using the primer pGL3-Basic/PromF. The HEK293 cell line was cultured as previously described (63). For analysis of luciferase activity, HEK293 cells were transfected by electroporation in OPTI-MEM I using Gene Pulser II (Bio-Rad) with 960 μF and 0.18 kV with 5 μg of pGL3-based reporter constructs, together with 150 ng of pRL-TKLuc as an internal control. Where indicated, cells were additionally transfected with 5 μg of PcDNA3-FLAG-JUNB, 5 μg of pcDNA-FLAG-BATF, or 40 μg of the respective pHEBO-IRF4 variants. Forty-eight hours after transfection, the ratio of the two luciferases was determined (dual luciferase kit; Promega).
Statistical analyses
For Figs. 1 to 6, SciPy (version 1.7.1) (65) was used for mul. Two-group data were compared using the Welch’s t test (one-tailed), and multiple-group data were compared using the Tukey’s honestly significant difference test. For figs. S1 to S9, Graphpad Prism (version 8) was used for statistical analysis. Multiple-group data were compared using one-way analysis of variance (ANOVA) and post hoc test, and two-group data were assessed by two-tailed unpaired Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. P values smaller than 0.05 are considered statistically significant. Error bars show the means ± SD.
Supplementary Material
Acknowledgments:
We thank the patients and their families for supporting the study. We acknowledge the extended clinical care team for supporting these patients, including the Rare Disease Discovery Hub at BC Children’s Hospital and for the support of this patient-centered research project. We thank the Bioinformatics Team of Fudan University Children’s Hospital, J. Qian and Y. Zhong at Shanghai Medical College Fudan University for excellent technical support, and the Animal Facility of Fudan University for maintaining the mice. We thank M. Führer, C. Kellerer, J. Nell, E. Rump, S. Wölfle, B. Winter, S. Omari, S. Lusatis, and B. Wollert-Wulf for excellent technical assistance, and H.-P. Rahn for cell sorting. We thank the Necker Imagine Centre de Resources Biologiques for generating EBV-transformed lymphoblastoid cell lines, the clinical research team at the Imagine Institute for the support, and the Etablissement Français du Sang for blood supply from healthy donors. We thank N.-C. Khin, N. Ross, and J. Lowe for assistance in generating the Irf4T9SR and Irf4 knockout mouse strains at ANU. S.K. is a Centre National de la Recherche Scientifique staff researcher. S.E.T. holds a Tier 1 Canada Research Chair in Pediatric Precision Health and the Aubrey J. Tingle Professor of Pediatric Immunology. H.S.K. and S.D.R. state that the content of this article does not necessarily reflect the views or policies of the U.S. Department of Health and Human Services nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Funding:
This work was supported by the following grants: National Natural Science Foundation of China (grant #91942302, #31870898, and #82011540008, to J.Y.W.), Ministry of Science and Technology of China (grant #2019YFE0100600 to J.Y.W.), Canadian Institutes for Health Research (grant #PJT-178054 to S.E.T. and grant #FDN-148403 to TRH), BC Children’s Hospital Foundation (to S.E.T.), Canada Research Chairs Program (to S.E.T.), National Institute Of Allergy And Infectious Diseases of the National Institutes of Health (grant #R21AI171466 to R.M.B.), National Institutes of Health, intramural research program, NIH Clinical Center and NIAID to S.R., Agence Nationale de la Recherche (grant #ANR-19-CE17-0012-01 to S.K. and ANR-19-CE17-0012-02 and ANR-19-CE17-0012-04), the French state (via the Agence Nationale de la Recherche’s “Investissments d’avenir” program (ANR-10-IAHU-01 to Institute Imagine), the Ligue Contre le Cancer–Comité de Paris, INSERM to S.K., the National Health and Medical Research Council of Australia (GNT2012498) to A.E., and by the Phenomics Translation Initiative, an Medical Research Future Funds funded program (#EPCD000035) and the National Collaborative Research Infrastructure Strategy (NCRIS) via Phenomics Australia. C.G. was supported by the German Research Foundation (grant no. 316249678–SFB 1279, subproject B05). O.F., G.N., and W.W.W. were supported by grants from the Canadian Institutes of Health Research (PJT-162120), Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (RGPIN-2017-06824), and the BC Children’s Hospital Foundation and Research Institute.
IRF4 International Consortium authors and contributions
Co-first authors (these authors contributed equally; listed alphabetically by last name). Oriol Fornes1, Alicia Jia2, Hye Sun Kuehn3, Qing Min4, Ulrich Pannicke5, Nikolai Schleussner6,7,8, Romane Thouenon9, Zhijia Yu10.
Clinician authors (listed alphabetically by last name).
María de los Angeles Astbury11, Catherine M. Biggs2, Miguel Galicchio12, Jorge Alberto Garcia-Campos13, Silvina Gismondi14, Guadalupe Gonzalez Villarreal15, Kyla J. Hildebrand2, Manfred Hönig16, Jia Hou4, Despina Moshous17,18, Stefania Pittaluga19, Xiaowen Qian20, Jacob Rozmus2, Ansgar S. Schulz16, Aidé Tamara Staines-Boone13, Bijun Sun4, Jinqiao Sun4, Schauer Uwe21, Edna Venegas-Montoya13, Wenjie Wang4, Xiaochuan Wang4, Wenjing Ying4, Xiaowen Zhai20, Qinhua Zhou4.
Scientist authors (listed alphabetically by last name).
Altuna Akalin22, Isabelle André9, Thomas F.E. Barth23, Bernd Baumann24, Anne Brüstle25, Gaetan Burgio25, Jacinta C. Bustamante26, Jean-Laurent Casanova27,28,29,30, Marco G. Casarotto31, Marina Cavazzana32, Loïc Chentout9, Ian A. Cockburn25, Mariantonia Costanza6,7,8, Chaoqun Cui33, Oliver Daumke34, Kate L. Del Bel2, Hermann Eibel35, Xiaoqian Feng33, Vedran Franke22, J Christof M. Gebhardt36, Andrea Götz5, Stephan Grunwald34, Bénédicte Hoareau37, Timothy R.Hughes38, Eva-Maria Jacobsen16, Martin Janz6,7,8, Arttu Jolma38, Chantal Lagresle-Peyrou32, Nannan Lai39, Yaxuan Li33, Susan Lin2, Henry Y. Lu2, Saul O. Lugo-Reyes40, Xin Meng33, Peter Möller23, Nidia Moreno-Corona9, Julie E. Niemela3, Gherman Novakovsky1, Jareb J. Perez-Caraballo41,42, Capucine picard43,44,45,46,47, Lucie Poggi9, Maria-Emilia Puig-Lombardi48, Katrina L. Randall25,49, Anja Reisser36, Yohann Schmitt50, Sandali Seneviratne25, Mehul Sharma2, Jennifer Stoddard3, Srinivasan Sundararaj31, Harry Sutton25, Linh Q. Tran41,42, Ying Wang4, Wyeth W. Wasserman1, Zichao Wen33, Wiebke Winkler6,7,8, Ermeng Xiong33, Ally W.H. Yang38, Meiping Yu4, Lumin Zhang39, Hai Zhang4, Qian Zhao51, Xin Zhen41,42.
Co-senior Co-corresponding authors (these authors contributed equally; listed alphabetically by last name).
Anselm Enders10, Sven Kracker9, Ruben Martinez-Barricarte41,42, Stephan Mathas6,7,8, Sergio D. Rosenzweig3, Klaus Schwarz5,52, Stuart E. Turvey2, Ji-Yang Wang4,33,53,54,55.
Affiliations
1Centre for Molecular Medicine and Therapeutics, Department of Medical Genetics, BC Children’s Hospital Research Institute, University of British Columbia, Vancouver, BC, Canada.2Department of Pediatrics, BC Children’s Hospital, University of British Columbia, Vancouver, BC, Canada.3Immunology Service, Department of Laboratory Medicine, Clinical Center, National Institutes of Health (NIH), Bethesda, MD, USA.4Department of Clinical Immunology, Children’s Hospital of Fudan University, National Children’s Medical Center, Shanghai, China.5Institute for Transfusion Medicine, University of Ulm, Ulm, Germany.6Charité–Universitätsmedizin Berlin, Hematology, Oncology and Tumor Immunology, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany.7Max-Delbrück-Center for Molecular Medicine in the Helmholtz Association (MDC), Group Biology of Malignant Lymphomas, Berlin, Germany.8Experimental and Clinical Research Center (ECRC), a cooperation between the Max-Delbrück-Center for Molecular Medicine in the Helmholtz Association and the Charité–Universitätsmedizin Berlin, Berlin, Germany.9Université Paris Cité, Laboratory of Human Lymphohematopoiesis, Imagine Institute, INSERM UMR 1163, Paris, France.10Centre for Personalised Immunology and Division of Immunology and Infectious Disease, John Curtin School of Medical Research, Australian National University, Canberra, ACT, Australia.11Infectología pediátrica, Hospital Español de Rosario, Rosario, Argentina.12Allergy and Immunology Service, Hospital de Niños VJ Vilela, Rosario, Argentina.13Immunology Service, Hospital de Especialidades Unidad Médica de Alta Especialidad (UMAE), 25 del Instituto Mexicano del Seguro Social (IMSS), Monterrey, México.14Servicio de Pediatría, Hospital Español de Rosario, Rosario, Argentina.15Hematology Service, Hospital de Especialidades Unidad Médica de Alta Especialidad (UMAE), 25 del Instituto Mexicano del Seguro Social (IMSS), Monterrey, México.16Department of Pediatrics and Adolescent Medicine, University Medical Center Ulm, Ulm, Germany.17Pediatric Immunology, Hematology and Rheumatology Department, Necker-Enfants Malades University Hospital, Assistance Publique Hôpitaux de Paris, Paris, France.18Laboratory “Genome Dynamics in the immune System”, INSERM UMR 1163, imagine Institute, Université Paris Cité, Paris, France.19Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD, USA.20Department of Hematology/Oncology, Children’s Hospital of Fudan University, Shanghai, China.21University Children’s Hospital, Ruhr University Bochum, Bochum, Germany.22Max-Deibrück-Center for Molecular Medicine in the Helmholtz Association (MDC), Bioinformatics and Omics Data Science Platform, Berlin institute for Medical Systems Biology, Berlin, Germany.23Department of Pathology, University Medical Center Ulm, Ulm, Germany.24Institute of Physiological Chemistry, University of Ulm, Ulm, Germany.25Division of Immunology and Infectious Disease, John Curtin School of Medical Research, Australian National University, Canberra, ACT, Australia.26Université de Paris Cité, Necker Hospital, Study Center for Primary Immunodeficiencies, Imagine Institute, Assistance publique des hôpitaux de Paris (APHP), Paris, France.27St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, Rockefeller University, New York, NY, USA.28Université Paris Cité, Laboratory of Human Genetics of infectious Diseases, Necker Branch INSERM U1163, Necker Hospital for Sick Children, Imagine institute, INSERM UMR 1163, Paris, France.29Pediatric Immunology-Hematology Unit, Necker Hospital for Sick Children, 75015, Paris, France.30Howard Hughes Medical Institute (HHMI), Rockefeller University, New York, NY, USA.31Research School of Biology, Australian National University, Canberra, ACT, Australia.32Université de Paris Cité, Necker Hospital, Biotherapy and Clinical investigation Centre-APHP, imagine institute, Paris, France.33Department of immunology, School of Basic Medical Sciences, Fudan University, Shanghai, China.34Max-Delbrück-Center for Molecular Medicine in the Helmholtz Association (MDC), Group Structural Biology, Berlin, Germany.35Department of Rheumatology and Clinical Immunology, Medical Center and Faculty of Medicine, University of Freiburg and Center for Chronic Immunodeficiency, Medial Center and Faculty of Medicine, University of Freiburg, Freiburg, Germany.36Department of Physics, Institute of Biophysics, University of Ulm, Ulm, Germany.37Plateforme de Cytométrie CyPS, UMS-37 PASS, Faculté de Médecine de Sorbonne Université, Paris, France.38Donnelly Centre, University of Toronto, Toronto, ON, Canada.39Key laboratory of Whole-Period Monitoring and Precise Intervention of Digestive Cancer, Shanghai Municipal Health Commission (SMHC), Minhang Hospital, Fudan University, Shanghai, China.40Immune Deficiencies Laboratory, National Institute of Pediatrics Secretariat of Health, Mexico City, México.41Division of Genetic Medicine, Department of Medicine, Vanderbilt Genetics Institute, Vanderbilt University Medical Center, Nashville, TN, USA.42Division of Molecular Pathogenesis, Department of Pathology, Microbiology, and Immunology, Vanderbilt Center for Immunobiology, Vanderbilt Institute for Infection, Immunology, and Inflammation, Vanderbilt University Medical Center, Nashville, TN, USA.43Université Paris Cité, Paris, France.44Study Center for Primary Immunodeficiencies, Necker-Enfants Malades Hospital, Assistance Publique Hôpitaux de Paris (APHP), Paris, France.45Laboratory of Lymphocyte Activation and Susceptibility to EBV Infection, Inserm UMR 1163, Institut Imagine, Paris, France.46Department of Pediatric Immunology, Hematology and Rheumatology, Necker-Enfants Malades Hospital, APHP, Paris, France.47Centre de références des déficits immunitaires Héréditaires (CEREDIH), Necker-Enfants Malades Hospital, APHP, Paris, France.48Paris-Descartes Bioinformatics Platform, Imagine Institute, Paris, France.49ANU Medical School, Australian National University, Canberra, ACT, Australia.50Genomics Core Facility, Institut Imagine-Structure Fédérative de Recherche Necker, INSERM U1163 et INSERM US24/CNRS UMS3633, Paris Descartes Sorbonne Paris Cite University, Paris, France.51Department of Neurology, Zhongshan Hospital, Fudan University, Shanghai, China.52Institute for Clinical Transfusion Medicine and Immunogenetics Ulm, German Red Cross Blood Transfusion Service Baden-Württemberg-Hessen and University Hospital Ulm, Ulm, Germany.53Department of Microbiology and Immunology, College of Basic Medical Sciences, Zhengzhou University, Zhengzhou, China.54Division of Biochemistry, Faculty of Pharmacy and Graduate School of Pharmaceutical Science, Keio University, Tokyo, Japan.55Shanghai Huashen Institute of Microbes and Infections, Shanghai, China.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The IRF4T95R mice are available from J.-Y.W. or A.E. upon request and are covered by a material transfer agreement with Fudan University or Australian National University, respectively. The ChIP-seq and RNA-seq data of EBV-B cells can be accessed in the Gene Expression Omnibus (GEO) repository under the accession numbers GSE199684 and GSE199685 (GSE199686 SuperSeries). The scRNA-seq data of human B cells and PBMC can be accessed in the GEO repository under the accession numbers GSE215936 and GSE215938. The RNA-seq data of Raji cells can be accessed by DOI: 10.5281/zenodo.7198935. The RNA-seq data of mouse splenic B cells can be accessed in the ArrayExpress under the ID: E-MTAB-12352. All other data needed to support the conclusions of the paper are present in the paper or the Supplementary Materials.
REFERENCES AND NOTES
- 1.Tangye SG, Al-Herz W, Bousfiha A, Cunningham-Rundles C, Franco JL, Holland SM, Klein C, Morio T, Oksenhendler E, Picard C, Puel A, Puck J, Seppänen MRJ, Somech R, Su HC, Sullivan KE, Torgerson TR, Meyts I, Human inborn errors of immunity: 2022 Update on the classification from the international union of immunological societies expert committee. J. Clin. Immunol 42, 1473–1507(2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyts I, Bousfiha A, Duff C, Singh S, Lau YL, Condino-Neto A, Bezrodnik L, Ali A, Adeli M, Drabwell J, Primary immunodeficiencies: A decade of progress and a promising future. Front. Immunol 11, 625753 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mittrücker HW, Matsuyama T, Grossman A, Kündig TM, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi PS, Mak TW, Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science 275, 540–543 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Tamura T, Yanai H, Savitsky D, Taniguchi T, The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol 26, 535–584 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Agnarelli A, Chevassut T, Mancini EJ, IRF4 in multiple myeloma-Biology, disease and therapeutic target. Leuk. Res 72, 52–58 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Acquaviva J, Chen X, Ren R, IRF-4 functions as a tumor suppressor in early B-cell development. Blood 112, 3798–3806 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R, Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol 7, 773–782 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Giasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, Khan AA, Ciofani M, Spooner CJ, Rutz S, Hackney J, Nurieva R, Escalante CR, Ouyang W, Littman DR, Murphy KM, Singh H, A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science 338, 975–980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sundararaj S, Seneviratne S, Williams SJ, Enders A, Casarotto MG, Structural determinants of the IRF4/DNA homodimeric complex. Nucleic Acids Res. 49, 2255–2265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J, Totoki Y, Chiba K, Sato-Otsubo A, Nagae G, Ishii R, Muto S, Kotani S, Watatani Y, Takeda J, Sanada M, Tanaka H, Suzuki H, Sato Y, Shiozawa Y, Yoshizato T, Yoshida K, Makishima H, Iwanaga M, Ma G, Nosaka K, Hishizawa M, Itonaga H, Imaizumi Y, Munakata W, Ogasawara H, Sato T, Sasai K, Muramoto K, Penova M, Kawaguchi T, Nakamura H, Hama N, Shide K, Kubuki Y, Hidaka T, Kameda T, Nakamaki T, Ishiyama K, Miyawaki S. S, Yoon S, Tobinai K, Miyazaki Y, Takaori-Kondo A, Matsuda F, Takeuchi K, Nureki O, Aburatani H, Watanabe T, Shibata T, Matsuoka M, Miyano S, Shimoda K, Ogawa S, Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat. Genet 47, 1304–1315 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Ochiai K, Maienschein-Cline M, Simonetti G, Chen J, Rosenthal R, Brink R, Chong AS, Klein U, Dinner AR, Singh H, Sciammas R, Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity 38, 918–929 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith T, Cunningham-Rundles C, Primary B-cell immunodeficiencies. Hum. Immunol 80 351–362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willis SN, Good-Jacobson KL, Curtis J, Light A, Tellier J, Shi W, Smyth GK, Tarlinton DM, Belz GT, Corcoran LM, Kallies A, Nutt SL, Transcription factor IRF4 regulates germinal center cell formation through a B cell-intrinsic mechanism. J. Immunol 192, 3200–3206 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Bollig N, Brüstle A, Kellner K, Ackermann W, Abass E, Raifer H, Camara B, Brendel C, Giel G, Bothur E, Huber M, Paul C, Elli A, Kroczek RA, Nurieva R, Dong C, Jacob R, Mak TW, Lohoff M, Transcription factor IRF4 determines germinal center formation through follicular T-helper cell differentiation. Proc. Natl. Acad. Sci. U.S.A 109, 8664–8669 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H, Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity 25, 225–236 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Brüstle A, Heink S, Huber M, Rosenplänter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, Lohoff M, The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat. Immunol 8, 958–966 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Raczkowski F, Ritter J, Heesch K, Schumacher V, Guralnik A, Höcker L, Raifer H, Klein M, Bopp T, Harb H, Kesper DA, Pfefferle PI, Grusdat M, Lang PA, Mittrücker HW, Huber M, The transcription factor Interferon Regulatory Factor 4 is required for the generation of protective effector CD8+T cells. Proc. Natl. Acad. Sci. U.S.A 110, 15019–15024 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, Zhang H, Shi X, Xiao X, Fan Y, Minze LJ, Wang J, Ghobrial RM, Xia J, Sciammas R, Li XC, Chen W, Ablation of transcription factor IRF4 promotes transplant acceptance by driving allogenic CD4+ T cell dysfunction. Immunity 47, 1114–1128.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyst MJ, Ekiert R, Guy J, Selfridge J, Koerner MV, Merusi C, De Sousa D, Bird A, Affinity for DNA contributes to NLS independent nuclear localization of MeCP2. Cell Rep. 24, 2213–2220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Escalante CR, Nistal-Villán E, Shen L, García-Sastre A, Aggarwal AK, Structure of IRF-3 bound to the PRDIII-I regulatory element of the human interferon-beta enhancer. Mol. Cell 26, 703–716 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Hammal F, de Langen P, Bergon A, Lopez F, Ballester B, ReMap 2022: A database of Human, mouse, Drosophila and Arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments. Nucleic Acids Res. 50, D316–D325 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, Kawli T, Davis CA, Dobin A, Kaul R, Halow J, Van Nostrand EL, Freese P, Gorkin DU, Shen Y, He Y, Mackiewicz M, Pauli-Behn F, Williams BA, Mortazavi A, Keller CA, Zhang XO, Elhajjajy SI, Huey J, Dickel DE, Snetkova V, Wei X, Wang X, Rivera-Mulia JC, Rozowsky J, Zhang J, Chhetri SB, Zhang J, Victorsen A, White KP, Visel A, Yeo GW, Burge CB, Lécuyer E, Gilbert DM, Dekker J, Rinn J, Mendenhall EM, Ecker JR, Kellis M, Klein RJ, Noble WS, Kundaje A, Guigó R, Farnham PJ, Cherry JM, Myers RM, Ren B, Graveley BR, Gerstein MB, Pennacchio LA, Snyder MP, Bernstein BE, Wold B, Hardison RC, Gingeras TR, Stamatoyannopoulos JA, Weng Z, Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novakovsky G, Fornes O, Saraswat M, Mostafavi S, Wasserman WW, ExplaiNN: Interpretable and transparent neural networks for genomics. bioRxiv 492818 (2022). 10.1101/2022.05.20.492818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ansel KM, Ngo VN, Hyman PL, Luther SA, Förster R, Sedgwick JD, Browning JL, Lipp M, Cyster JG, A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature 406, 309–314 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Muller HJ, Further studies on the nature and causes of gene mutations. Proc. Sixth Int. Congr. Genet. 1, 213–255 (1932). [Google Scholar]
- 26.Arruabarrena-Aristorena A, Maag JLV, Kittane S, Cai Y, Karthaus WR, Ladewig E, Park J, Kannan S, Ferrando L, Cocco E, Ho SY, Tan DS, Sallaku M, Wu F, Acevedo B, Selenica P, Ross DS, Witkin M, Sawyers CL, Reis-Filho JS, Verma CS, Jauch R, Koche R, Baselga J, Razavi P, Toska E, Scaltriti M, FOXA1 mutations reveal distinct chromatin profiles and influence therapeutic response in breast cancer. Cancer Cell 38, 534–550.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia YK, Zeng YR, Zhang ML, Liu P, Liu F, Zhang H, He CX, Sun YP, Zhang JY, Zhang C, Song L, Ding C, Tang YJ, Yang Z, Yang C, Wang P, Guan KL, Xiong Y, Ye D, Tumor-derived neomorphic mutations in ASXL1 impairs the BAP1-ASXL1-FOXK1/K2 transcription network. Protein Cell 12, 557–577 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun Z, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Löwenberg B, Licht JD, Godley LA, Delwel R, Valk PJ, Thompson CB, Levine RL, Melnick A, Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lui JC, Raimann A, Hojo H, Dong L, Roschger P, Kikani B, Wintergerst U, Fratzl-Zelman N, Jee YH, Haeusler G, Baron J, A neomorphic variant in SP7 alters sequence specificity and causes a high-turnover bone disorder. Nat. Commun 13, 700 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Planutis A, Xue L, Trainor CD, Dangeti M, Gillinder K, Siatecka M, Nebor D, Peters LL, Perkins AC, Bieker JJ, Neomorphic effects of the neonatal anemia (Nan-Eklf) mutation contribute to deficits throughout development. Development 144, 430–440 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pippucci T, Maresca A, Magini P, Cenacchi G, Donadio V, Palombo F, Papa V, Incensi A, Gasparre G, Valentino ML, Preziuso C, Pisano A, Ragno M, Liguori R, Giordano C, Tonon C, Lodi R, Parmeggiani A, Carelli V, Seri M, Homozygous NOTCH3 null mutation and impaired NOTCH3 signaling in recessive early-onset arteriopathy and cavitating leukoencephalopathy. EMBO Mol. Med 7, 848–858(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guérin A, Kerner G, Marr N, Markle JG, Fenollar F, Wong N, Boughorbel S, Avery DT, Ma CS, Bougarn S, Bouaziz M, Béziat V, Mina ED, Oleaga-Quintas C, Lazarov T, Worley L, Nguyen T, Patin E, Deswarte C, Martinez-Barricarte R, Boucherit S, Ayral X, Edouard S, Boisson-Dupuis S, Rattina V, Bigio B, Vogt G, Geissmann F, Quintana-Murci L, Chaussabel D, Tangye SG, Raoult D, Abel L, Bustamante J, Casanova JL, IRF4 haploinsufficiency in a family with Whipple's disease. eLife 7, e32340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.García-Morato MB, Santos FJA, Briones AC, Moreno AB, Maté ÁDP, Domínguez-Soto A, Merino MJB, Molina LDP, Canizales JT, Marin AV, García EV, Rodríguez MF, Sabando DPL, Jiménez-Reinoso A, Del Castillo YM, Santaeufemia FJS, de Lucas-Laguna R, Cárdenas PP, Polo LC, Díaz MC, Valés-Gómez M, Santiago ER, Cerdán AF, Blanco JN, Corbí ÁL, Reyburn HT, Regueiro JR, López-Granados E, Pena RR, New human combined immunodeficiency caused by interferon regulatory factor 4 (IRF4) deficiency inherited by uniparental isodisomy. J. Allergy Clin. Immunol 141, 1924–1927.e18 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Huber M, Lohoff M, IRF4 at the crossroads of effector T-cell fate decision. Eur. J. Immunol 44, 1886–1895 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Field MA, Cho V, Andrews TD, Goodnow CC, Reliably detecting clinically important variants requires both combined variant calls and optimized filtering strategies. PLOS ONE 10, e0143199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai N, Liu L, Lin L, Cui C, Wang Y, Min Q, Xiong E, Wang W, Ying W, Zhou Q, Hou J, Sun J, Wang JY, Wang X, Effective and safe treatment of a novel IL2RA deficiency with rapamycin. J. Allergy Clin. Immunol. Pract 8, 1132–1135.e4 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Liu X, Li C, Mou C, Dong Y, Tu Y, dbNSFP v4: A comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 12, 103 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sic H, Kraus H, Madl J, Flittner KA, von Münchow AL, Pieper K, Rizzi M, Kienzler AK, Ayata K, Rauer S, Kleuser B, Salzer U, Burger M, Zirlik K, Lougaris V, Plebani A, Römer W, Loeffler C, Scaramuzza S, Villa A, Noguchi E, Grimbacher B, Eibel H, Sphingosine-1-phosphate receptors control B-cell migration through signaling components associated with primary immunodeficiencies, chronic lymphocytic leukemia, and multiple sclerosis. J. Allergy Clin. Immunol 134, 420–428(2014). [DOI] [PubMed] [Google Scholar]
- 39.Smulski CR, Kury P, Seidel LM, Staiger HS, Edinger AK, Willen L, Seidl M, Hess H, Salzer U, Rolink AG, Rizzi M, Schneider P, Eibel H, BAFF-and TACI-dependent processing of BAFFR by ADAM proteases regulates the survival of B cells. Cell Rep. 18, 2189–2202 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Sevdali E, Block V, Lataretu M, Li H, Smulski CR, Briem JS, Heitz Y, Fischer B, Ramirez NJ, Grimbacher B, Jäck HM, Voll RE, Hölzer M, Schneider P, Eibel H, BAFFR activates PI3K/AKT signaling in human naive but not in switched memory B cells through direct interactions with B cell antigen receptors. Cell Rep. 39, 111019 (2022). [DOI] [PubMed] [Google Scholar]
- 41.Smulski CR, Zhang L, Burek M, Teixidó Rubio A, Briem JS, Sica MP, Sevdali E, Vigolo M, Willen L, Odermatt P, Istanbullu D, Herr S, Cavallari M, Hess H, Rizzi M, Eibel H, Schneider P, Ligand-independent oligomerization of TACI is controlled by the transmembrane domain and regulates proliferation of activated B cells. Cell Rep. 38, 110583 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Locci M, Wu JE, Arumemi F, Mikulski Z, Dahlberg C, Miller AT, Crotty S, Activin A programs the differentiation of human TFH cells. Nat. Immunol 17, 976–984 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tosato G, Cohen JI, Generation of Epstein-Barr Virus (EBV)-immortalized B cell lines. Curr. Protoc. Immunol Chapter 7, Unit 7.22 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Roe CE, Hayes MJ, Barone SM, Irish JM, Training novices in generation and analysis of high-dimensional human cell phospho-flow cytometry data. Curr. Protoc. Cytom 93, e71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nowicka M, Krieg C, Crowell HL, Weber LM, Hartmann FJ, Guglietta S, Becher B, Levesque MP, Robinson MD, CyTOF workflow:Differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res 6, 748(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, Saeys Y, FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Diggins KE, Greenplate AR, Leelatian N, Wogsland CE, Irish JM, Characterizing cell subsets using marker enrichment modeling. Nat. Methods 14, 275–278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pannicke U, Hönig M, Hess I, Friesen C, Holzmann K, Rump EM, Barth TF, Rojewski MT, Schulz A, Boehm T, Friedrich W, Schwarz K, Reticular dysgenesis (aleukocytosis) is caused by mutations in the gene encoding mitochondrial adenylate kinase 2. Nat. Genet 41, 101–105 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Xiong E, Li Y, Min Q, Cui C, Liu J, Hong R, Lai N, Wang Y, Sun J, Matsumoto R, Takahashi D, Hase K, Shinkura R, Tsubata T, Wang JY, MZB1 promotes the secretion of J-chain-containing dimeric IgA and is critical for the suppression of gut inflammation. Proc. Natl. Acad. Sci. U.S.A 116, 13480–13489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Randall KL, Lambe T, Johnson AL, Treanor B, Kucharska E, Domaschenz H, Whittle B, Tze LE, Enders A, Crockford TL, Bouriez-Jones T, Alston D, Cyster JG, Lenardo MJ, Mackay F Deenick EK, Tangye SG, Chan TD, Camidge T, Brink R, Vinuesa CG, Batista FD, Cornall RJ, Goodnow CC, Dock8 mutations cripple B cell immunological synapses, germinal centers and long-lived antibody production. Nat. Immunol 10, 1283–1291 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Espinosa DA, Christensen D, Muñoz C, Singh S, Locke E, Andersen P, Zavala F, Robust antibody and CD8+ T-cell responses induced by P. falciparum CSP adsorbed to cationic liposomal adjuvant CAF09 confer sterilizing immunity against experimental rodent malaria infection. NPJ Vaccines 2, 10(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J, Xiong E, Zhu H, Mori H, Yasuda S, Kinoshita K, Tsubata T, Wang JY, Efficient induction of Ig gene hypermutation in ex vivo-activated primary B cells. J. Immunol 199, 3023–3030 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Wurmus R, Uyar B, Osberg B, Franke V, Gosdschan A, Wreczycka K, Ronen J, Akalin A, PiGx: Reproducible genomics analysis pipelines with GNU Guix. Gigascience 7, giyl 23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrel I D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, Gil L, Gordon L, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, To JK, Laird MR, Lavidas I, Liu Z, Loveland JE, Maurel T, McLaren W, Moore B, Mudge J, Murphy DN, Newman V, Nuhn M, Ogeh D, Ong CK, Parker A, Patricio M, Riat HS, Schuilenburg H, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Zadissa A, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Cunningham F, Yates A, Flicek P, Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C, Salmon provides fast and biasaware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soneson C, Love MI, Robinson MD, Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biot. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu Z, Eils R, Schlesner M, Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Lachmann A, Torre D, Keenan AB, Jagodnik KM, Lee HJ, Wang L, Silverstein MC, Ma'ayan A, Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun 9, 1366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng L, Liu J, Niu L, Kamran M, Yang AWH, Jolma A, Dai Q, Hughes TR, Patel DJ, Zhang L, Prasanth SG, Yu Y, Ren A, Lai EC, Distinct structural bases for sequence-specific DNA binding by mammalian BEN domain proteins. Genes Dev. 36, 225–240 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marçais G, Kingsford C, A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770(2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Rob- bertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O'Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD, Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kreher S, Bouhlel MA, Cauchy P, Lamprecht B, Li S, Grau M, Hummel F, Köchert K, Anagnostopoulos I, Jöhrens K, Hummel M, Hiscott J, Wenzel SS, Lenz P, Schneider M, Küppers R, Scheidereit C, Giefing M, Siebert R, Rajewsky K, Lenz G, Cockerill PN, Janz M, Dörken B, Bonifer C, Mathas S, Mapping of transcription factor motifs in active chromatin identifies IRF5 as key regulator in classical Hodgkin lymphoma. Proc. Natl. Acad. Sci. USA 111, E4513–E4522 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mathas S, Janz M, Hummel F, Hummel M, Wollert-Wulf B, Lusatis S, Anagnostopoulos I, Lietz A, Sigvardsson M, Jundt F, Jøhrens K, Bommert K, Stein H, Dörken B, Intrinsic inhibition of transcription factor E2A by HLH proteins ABF-1 and Id2 mediates reprogramming of neoplastic B cells in Hodgkin lymphoma. Nat. Immunol 7, 207–215 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, van der Walt SJ, Brett M, Wilson J, Millman KJ, Mayorov N, Nelson ARJ, Jones E, Kern R, Larson E, Carey CJ, Polat I, Feng Y, Moore EW, VanderPlas J, Laxalde D, Perktold J, Cimrman R, Henriksen I, Quintero EA, Harris CR, Archibald AM, Ribeiro AH, Pedregosa F, van Mulbregt P, SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Min Q, Meng X, Zhou Q, Wang Y, Li Y, Lai N, Xiong E, Wang W, Yasuda S, Yu M, Zhang H, Sun J, Wang X, Wang JY, RAG1 splicing mutation causes enhanced B cell differentiation and autoantibody production. JCI Insight 6, e148887 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morita S, Kojima T, Kitamura T, Plat-E: An efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066(2000). [DOI] [PubMed] [Google Scholar]
- 68.Hipp L, Beer J, Kuchler O, Reisser M, Sinske D, Michaelis J, Gebhardt JCM, Knöll B, Single-molecule imaging of the transcription factor SRF reveals prolonged chromatinbinding kinetics upon cell stimulation. Proc. Natl. Acad. Sci. U.S.A 116, 880–889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reisser M, Palmer A, Popp AP, Jahn C, Weidinger G, Gebhardt JCM, Single-molecule imaging correlates decreasing nuclear volume with increasing TF-chromatin associations during zebrafish development. Nat. Commun 9, 5218(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Popp AP, Hettich J, Gebhardt JCM, Altering transcription factor binding reveals comprehensive transcriptional kinetics of a basic gene. Nucleic Acids Res. 49, 6249–6266 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuhn T, Hettich J, Davtyan R, Gebhardt JCM, Single molecule tracking and analysis framework including theory-predicted parameter settings. Sci. Rep 11, 9465 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koo PK, Ploenzke M, Improving representations of genomic sequence motifs in convolutional networks with exponential activations. Nat. Mach. Intell 3, 258–266 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quinlan AR, Hall IM, BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khan A, Puig RR, Boddie P, Mathelier A, BiasAway: Command-line and web server to generate nucleotide composition-matched DNA background sequences. Bioinformatics 37, 1607–1609 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E, Louppe G, Scikit-learn: Machine learning in python. J. Mach. Learn. Res 12, 2825–2830 (2012). [Google Scholar]
- 76.Asif M, Orenstein Y, DeepSELEX: Inferring DNA-binding preferences from HT-SELEX data using multi-class CNNs. Bioinformatics 36, i634–i642 (2020). [DOI] [PubMed] [Google Scholar]
- 77.Kingma DP, Ba JJC, Adam: A method for stochastic optimization. arXiv:1412.6980 [cs.LG] (2015). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.