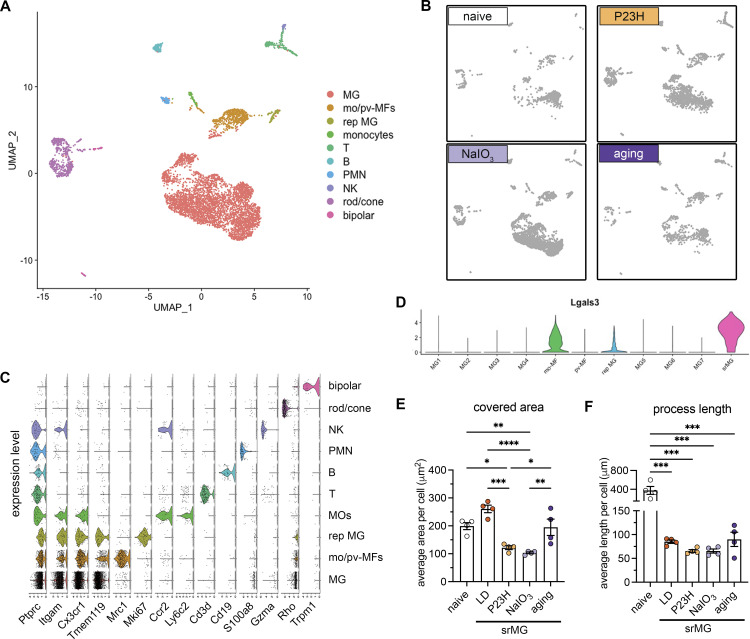
Figure S1.
scRNA-seq and morphological analysis of subretinal microglia across mouse models of outer retinal degeneration. (A and B) UMAP plots showing retinal CD45+ cells collected from naïve mice, NaIO3 mediated RPE injury model, P23H model, and advanced aging model as indicated. MG, microglia; mo-MFs, monocyte-derived macrophages; pv-MFs: perivascular macrophages; mo-DCs, monocyte-derived dendritic cells; PMN, polymorphonuclear neutrophils; NK, natural killer. (C) Violin plots show marker expressions for each cluster. (D) Violin plots showing Lgals3 expression across all macrophage clusters. srMG, subretinal microglia. (E and F) Quantifications of covered area and process length in naïve microglia from the inner retina and subretinal microglia from four mouse models of retinal degenerations (n = 4 mice per group). Data were collected from two independent experiments for sequencing and validation. *: P < 0.05; **: P < 0.01; ***: P < 0.001; ****: P < 0.0001 (one-way ANOVA with Tukey’s post hoc test).